Feature #1861
Implement multiplicative spectral models
| Status: | Closed | Start date: | 10/03/2016 | ||
|---|---|---|---|---|---|
| Priority: | Normal | Due date: | |||
| Assigned To: | % Done: | 90% | |||
| Category: | - | ||||
| Target version: | 1.2.0 | ||||
| Duration: |
Description
To implement for example EBL absorption, multiplicative spectral model should be implemented. After discussion at today’s coding sprint it appears best to implement the multiplicative spectral model in analogy to the composite spectral models (see #1706). A possible XML format for the model definition would be:
<?xml version="1.0" standalone="no"?>
<source_library title="source library">
<source name="Crab" type="PointSource">
<spatialModel type="PointSource">
<parameter name="RA" scale="1.0" value="83.6331" min="-360" max="360" free="1"/>
<parameter name="DEC" scale="1.0" value="22.0145" min="-90" max="90" free="1"/>
</spatialModel>
<spectrum type="Multiplicative">
<spectrum type="PowerLaw">
<parameter name="Prefactor" scale="1e-16" value="5.7" min="1e-07" max="1000.0" free="1"/>
<parameter name="Index" scale="-1" value="2.48" min="0.0" max="+5.0" free="1"/>
<parameter name="PivotEnergy" scale="1e6" value="0.3" min="0.01" max="1000.0" free="0"/>
</spectrum>
<spectrum type="ExponentialCutoffPowerLaw">
<parameter name="Prefactor" scale="1.0" value="1.0" min="1e-07" max="1000.0" free="0"/>
<parameter name="Index" scale="1.0" value="0.0" min="-2.0" max="+2.0" free="0"/>
<parameter name="CutoffEnergy" scale="1e6" value="1.0" min="0.01" max="1000.0" free="1"/>
<parameter name="PivotEnergy" scale="1e6" value="0.3" min="0.01" max="1000.0" free="0"/>
</spectrum>
</spectrum>
</source>
</source_library>
GModelSpectralMultiplicative class.
Recurrence
No recurrence.
History
#1
 Updated by Mayer Michael over 8 years ago
Updated by Mayer Michael over 8 years ago
- Status changed from New to In Progress
- Assigned To set to Mayer Michael
- Target version set to 1.2.0
- % Done changed from 0 to 80
I have implemented the multiplicative model on branch 1861-multiplicative-spectral-model.
I am about to start the pull distributions.
#2
 Updated by Mayer Michael over 8 years ago
Updated by Mayer Michael over 8 years ago
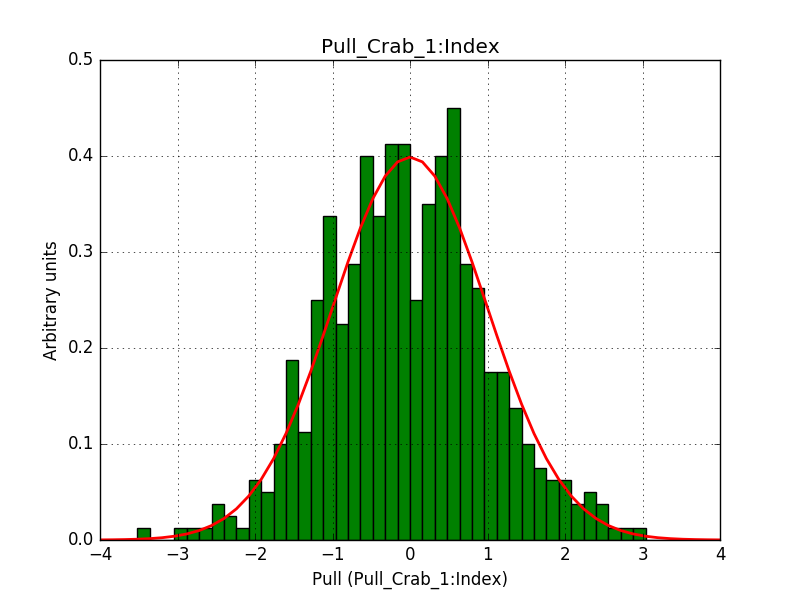
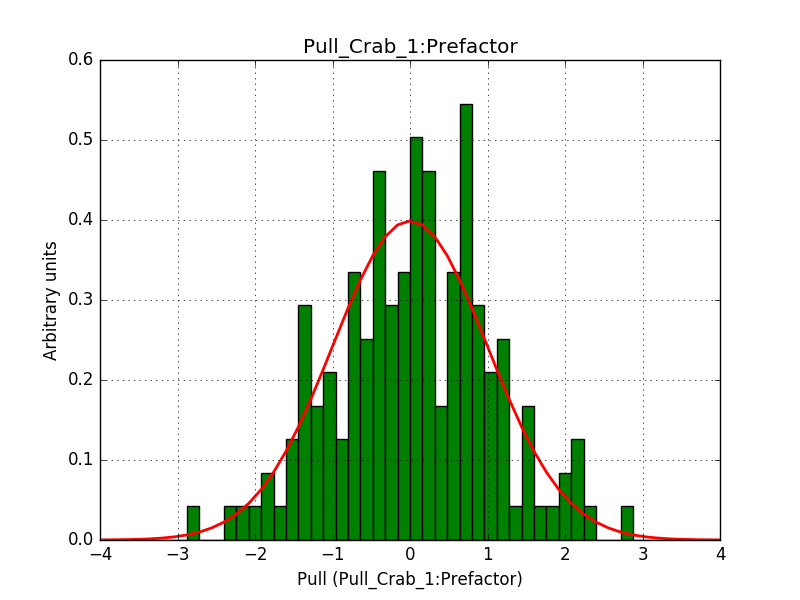
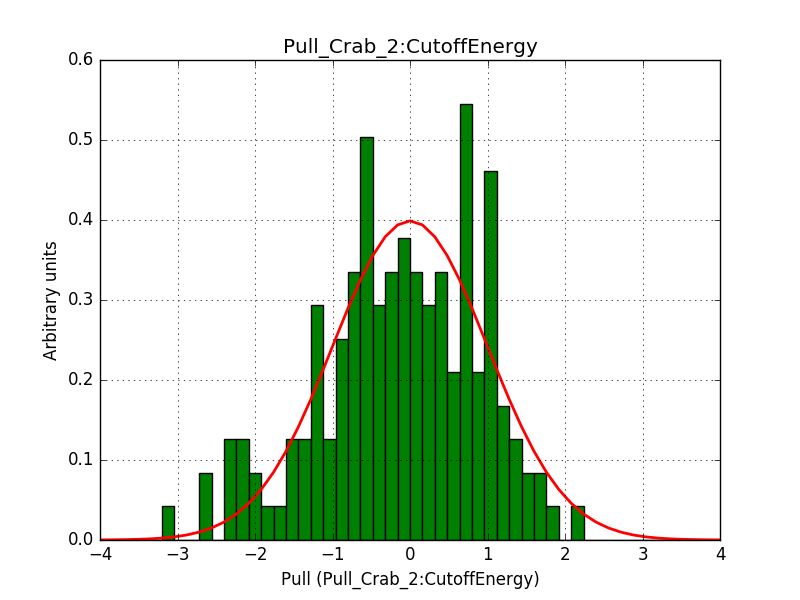
- File 1_Index.png added
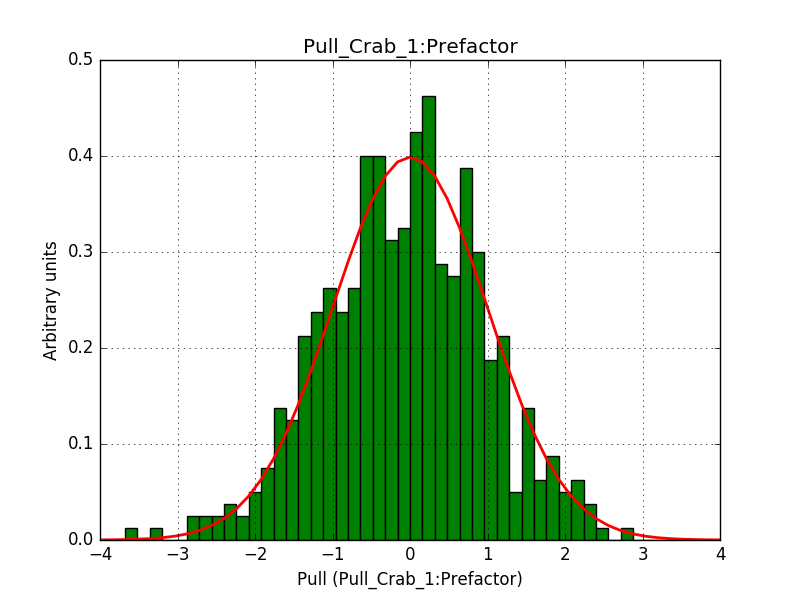
- File 1_Prefactor.png added
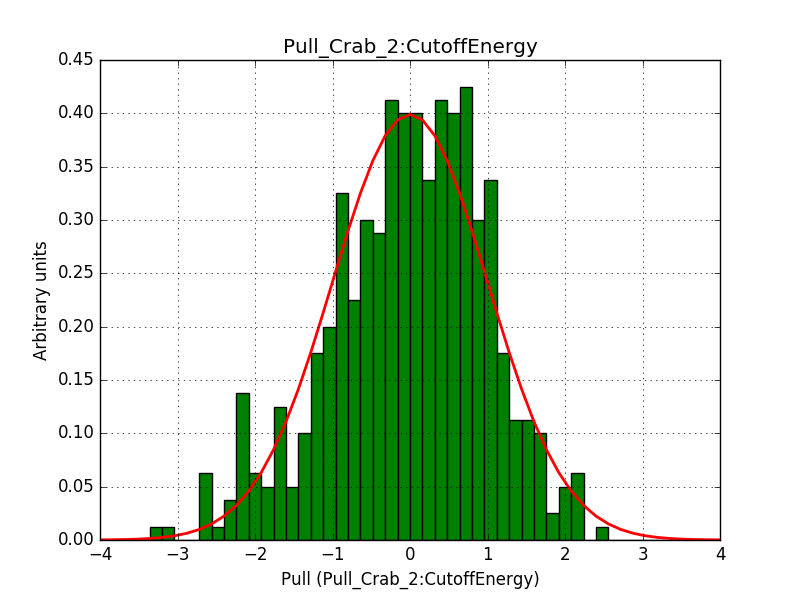
- File 2_CutoffEnergy.png added
- Status changed from In Progress to Pull request
- % Done changed from 80 to 100
Below are the pull distributions with 500 trials using the above model including a CTA background model. User documentation is still missing, but we could move this to a separate feature?.
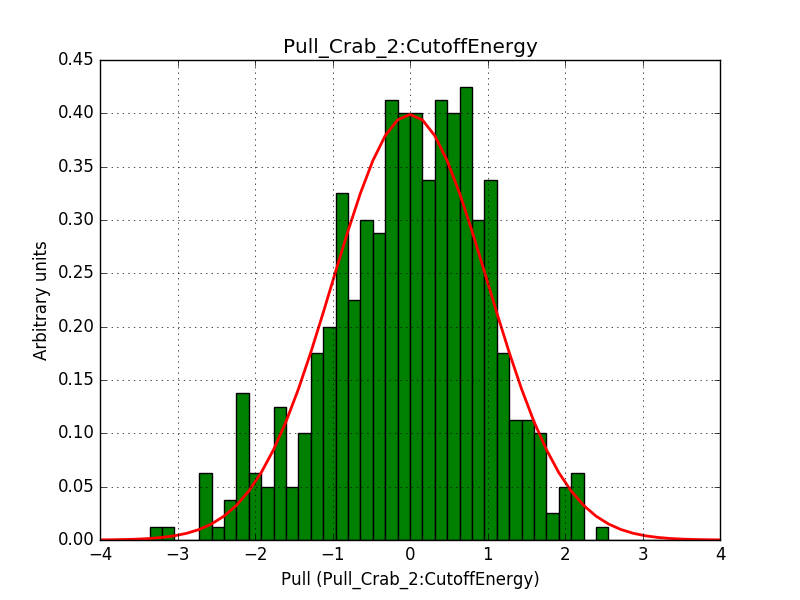
#3
 Updated by Mayer Michael over 8 years ago
Updated by Mayer Michael over 8 years ago
I added related feature #1865 to not forget about documentation 
#4
 Updated by Knödlseder Jürgen over 8 years ago
Updated by Knödlseder Jürgen over 8 years ago
- Status changed from Pull request to In Progress
- % Done changed from 100 to 90
I merged code into devel.
I added some code so that the update method for Monte Carlo also checks whether any parameters have been changed. It would be good to rerun the pull distribution test to see whether the code still works fine.
Would also be good to add a minimum amount of documentation. At least on the ctools user side it would be nice to have an explanation of how to use multiplicative spectral models.
I also recognised that the formula for the gradients is missing in the eval() header. Would be good to write it down.
Keep this feature open for the moment so that we are reminded of these remaining issues (well, at least the additional checking and formula for gradients 
#5
 Updated by Mayer Michael over 8 years ago
Updated by Mayer Michael over 8 years ago
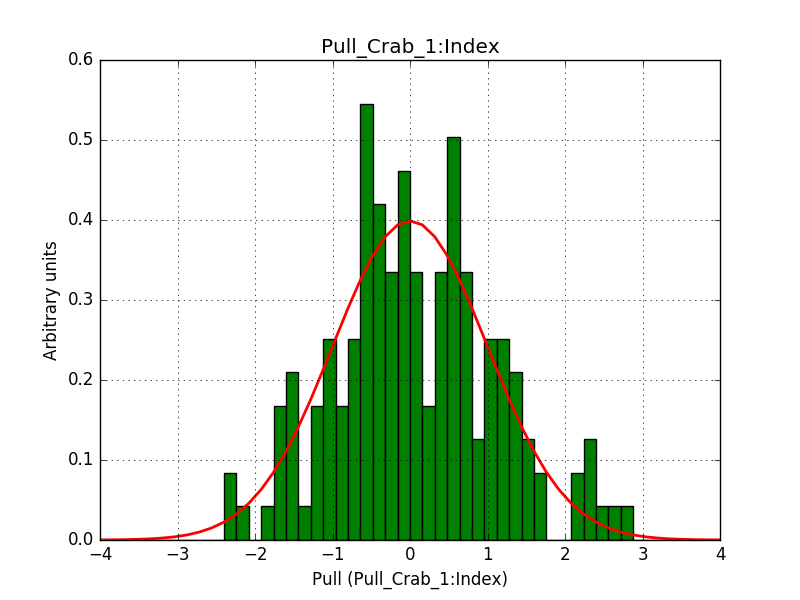
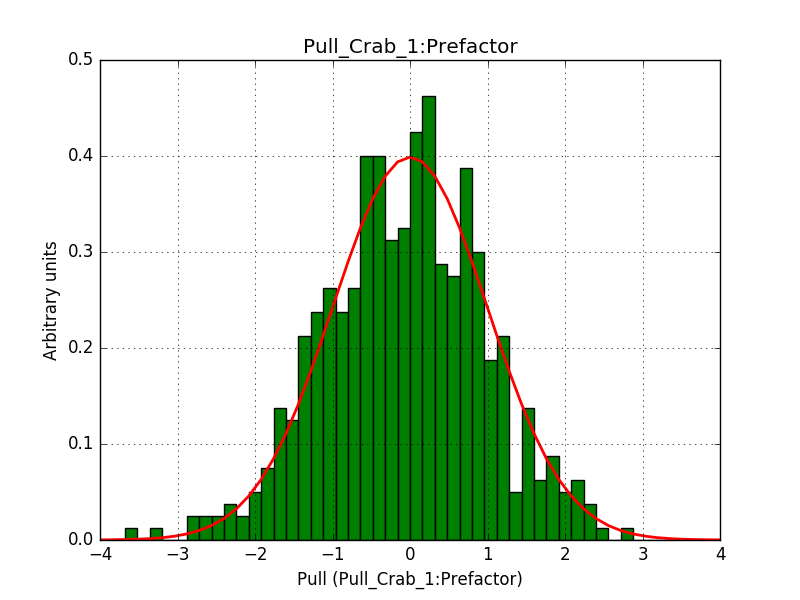
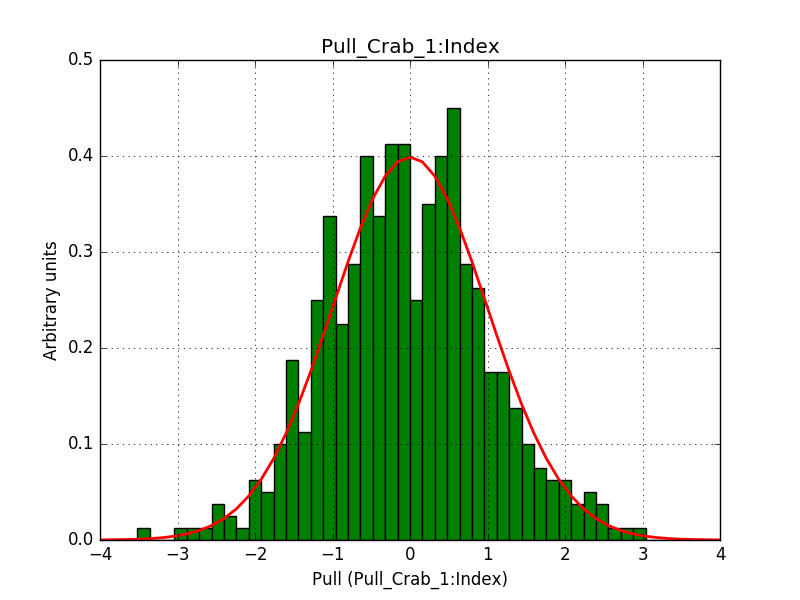
- File 1_Index_2.png added
- File 1_Prefactor_2.png added
- File 2_CutoffEnergy_2.png added
- Status changed from In Progress to Pull request
I have added the LaTeX formula of the gradients to the doxygen documentation. Furthermore I removed the add_component() method since it was not needed and could be merged in the read method. A check for unique component names has also been implemented and a corresponding function was added to GTools.
For the documentation, I added #1865.
I forgot to work on my old branch. For simplicity I now created a new branch where I made the changes: 1861-multiplicative-spectral-model-continued
Pull distributions have started and look good. Here is a snapshot of the distributions of the first 150 trials.
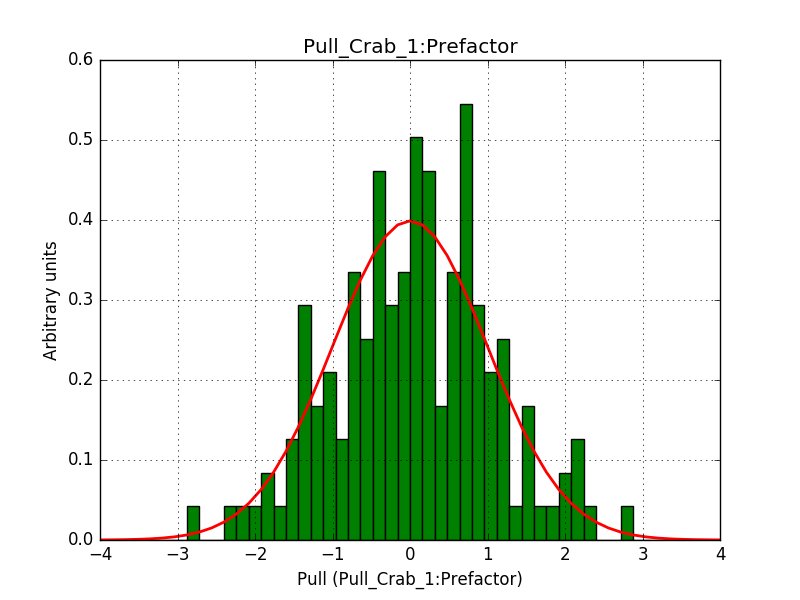
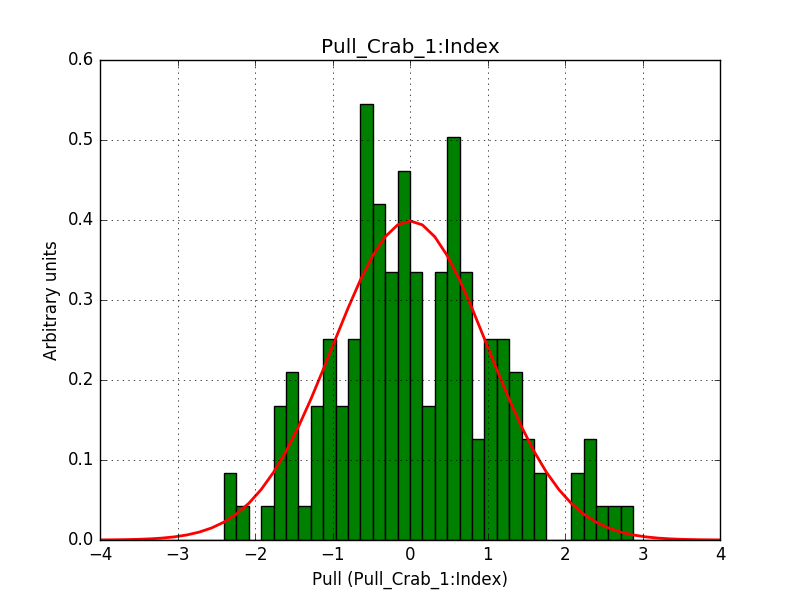
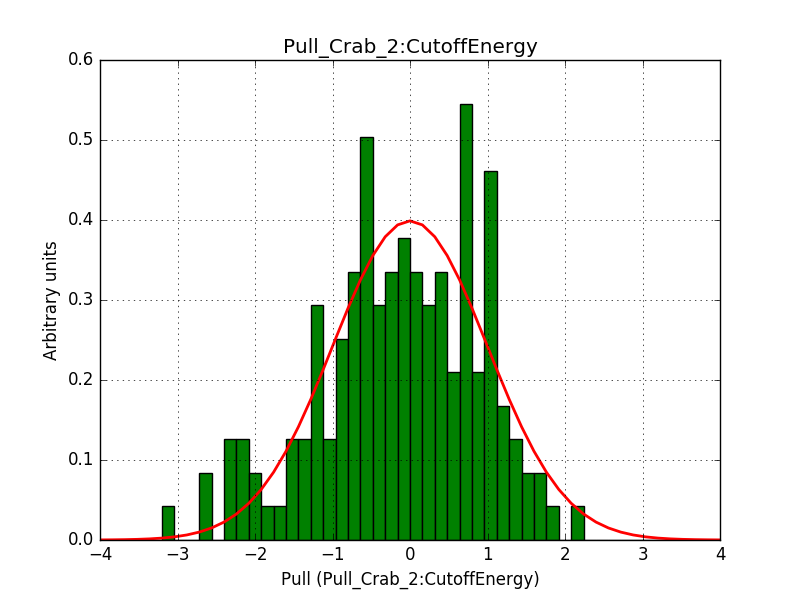
#6
 Updated by Knödlseder Jürgen over 8 years ago
Updated by Knödlseder Jürgen over 8 years ago
- Status changed from Pull request to Closed
Merged into devel.