Feature #1914
Add GModelTemporalFunc class
| Status: | Closed | Start date: | 01/24/2017 | |
|---|---|---|---|---|
| Priority: | Normal | Due date: | ||
| Assigned To: | % Done: | 100% | ||
| Category: | - | |||
| Target version: | 1.2.0 | |||
| Duration: |
Description
The GModelTemporalFunc should implement a temporal file function model, comprised of nodes that define the relative intensity at given times. The file function should be defined in a FITS file, including the definition of a time reference so that the value of the time column is defined in absolute times. The unit of the time column should be seconds, the file function should be unit less.
Recurrence
No recurrence.
History
#1
 Updated by Knödlseder Jürgen about 8 years ago
Updated by Knödlseder Jürgen about 8 years ago
- File validate1.png added
- File validate2.png added
- File validate3.png added
- File validate4.png added
- File validate5.png added
- File validate6.png added
- File test-filefunction.py
 added
added - File temp_filefunction.fits added
- Target version set to 1.2.0
- % Done changed from 0 to 90
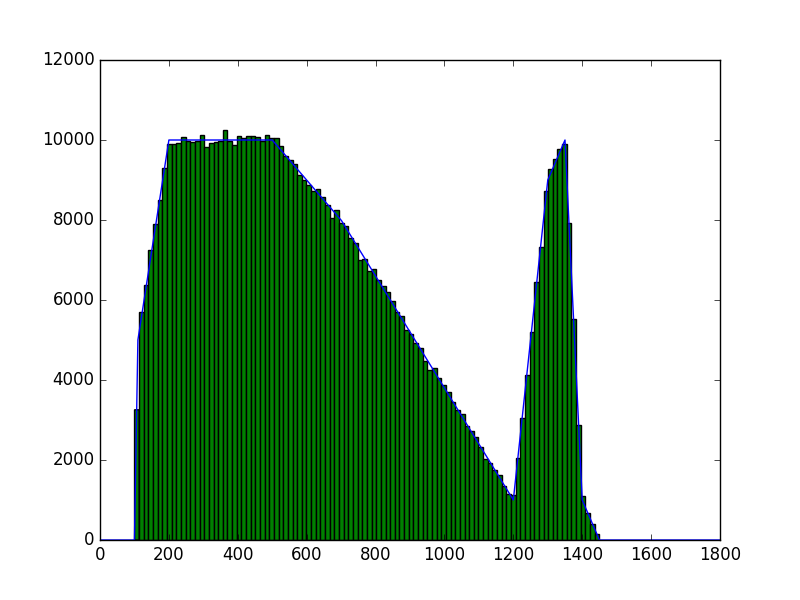
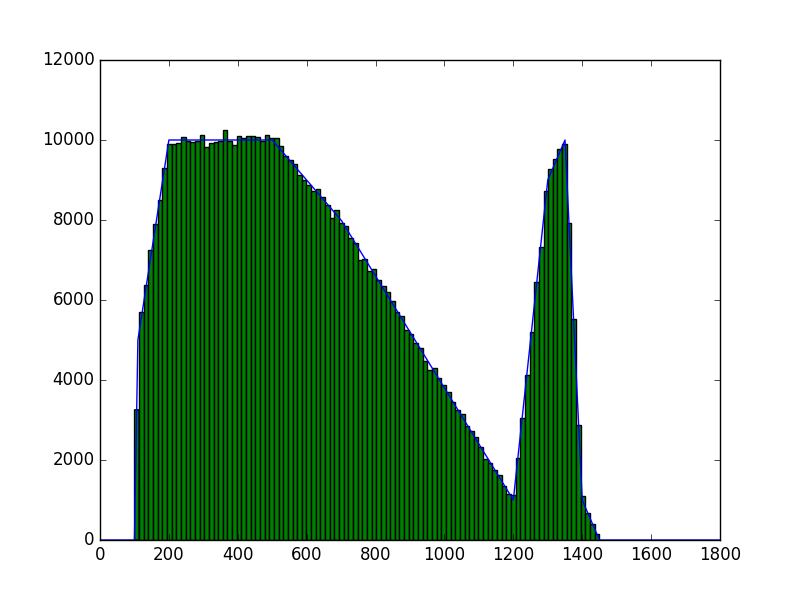
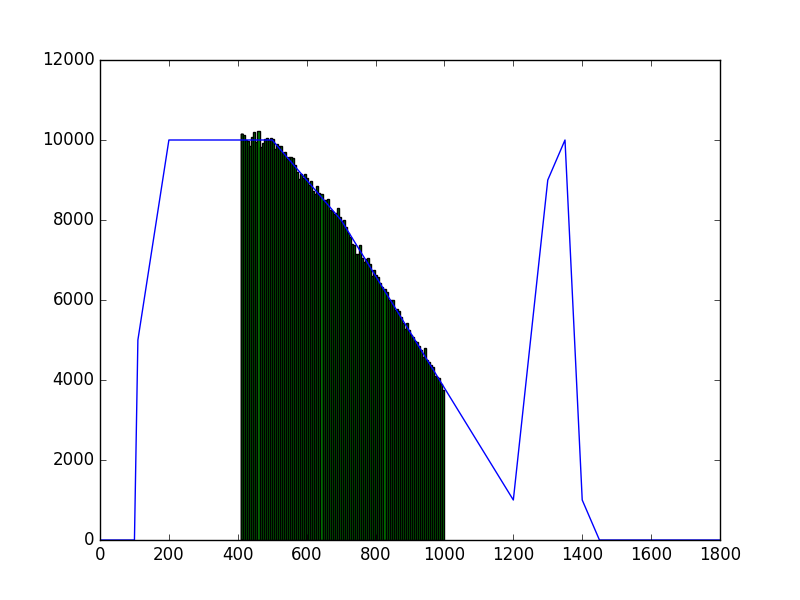
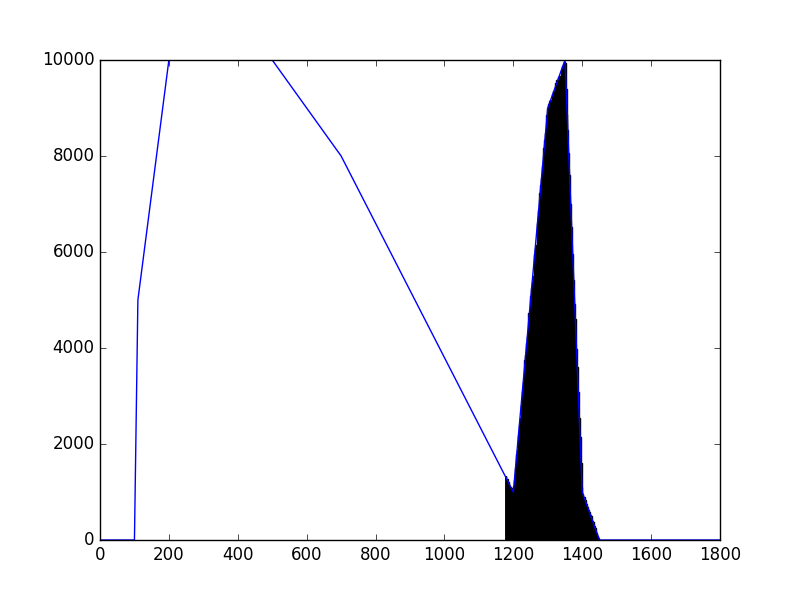
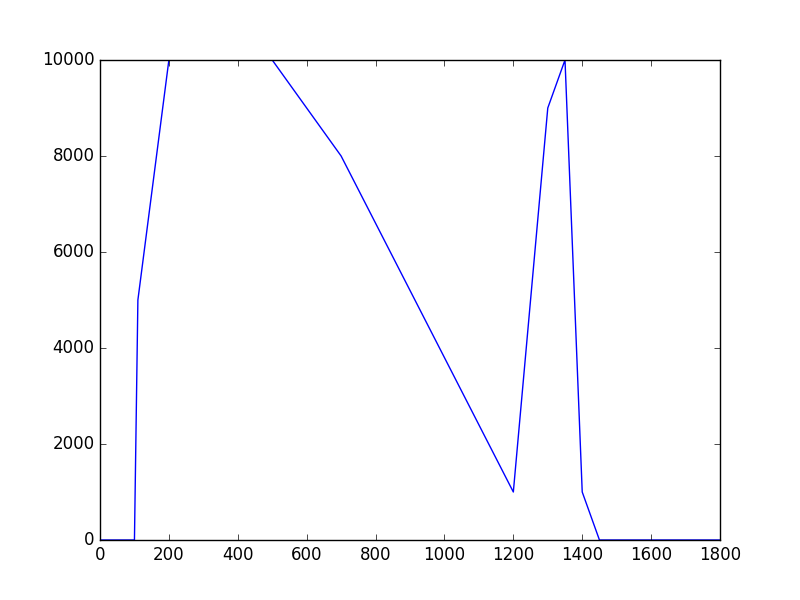
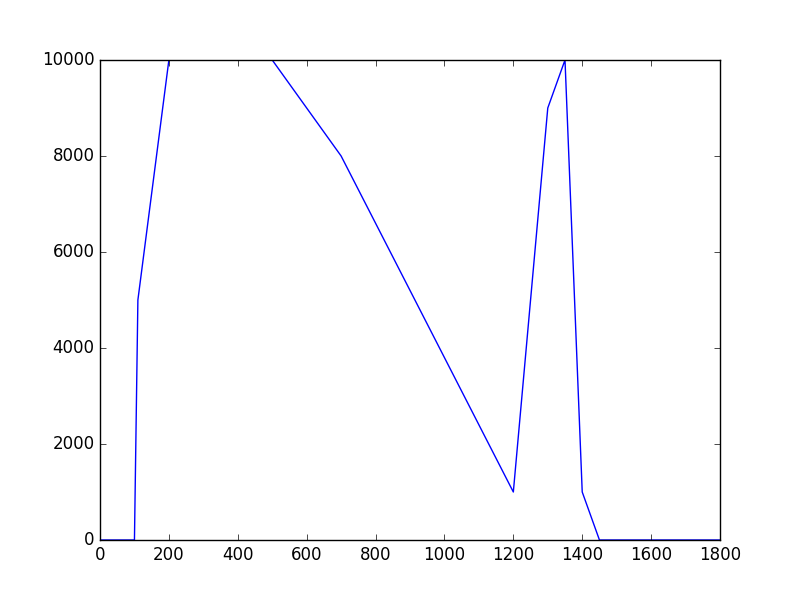
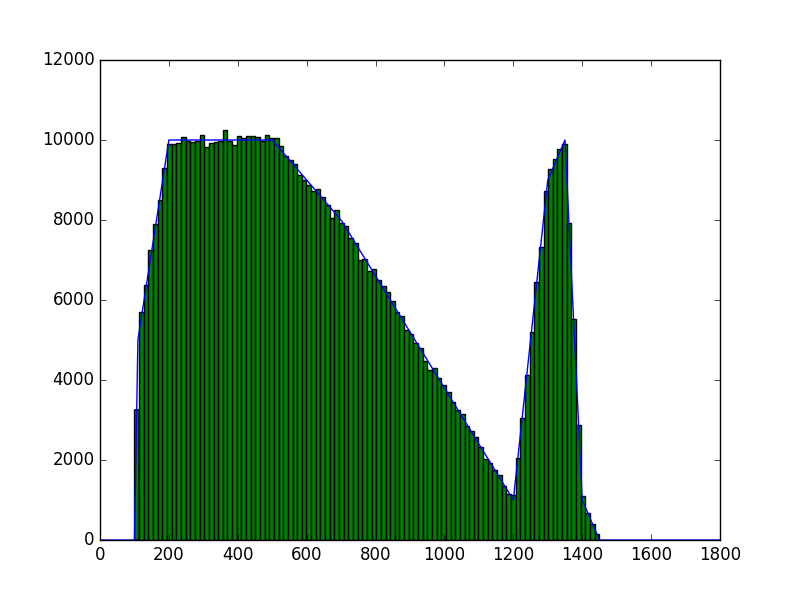
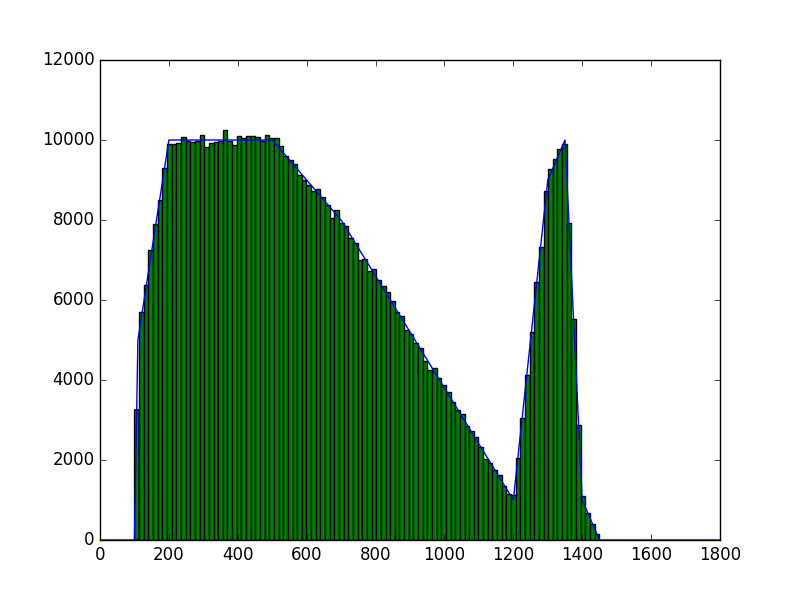
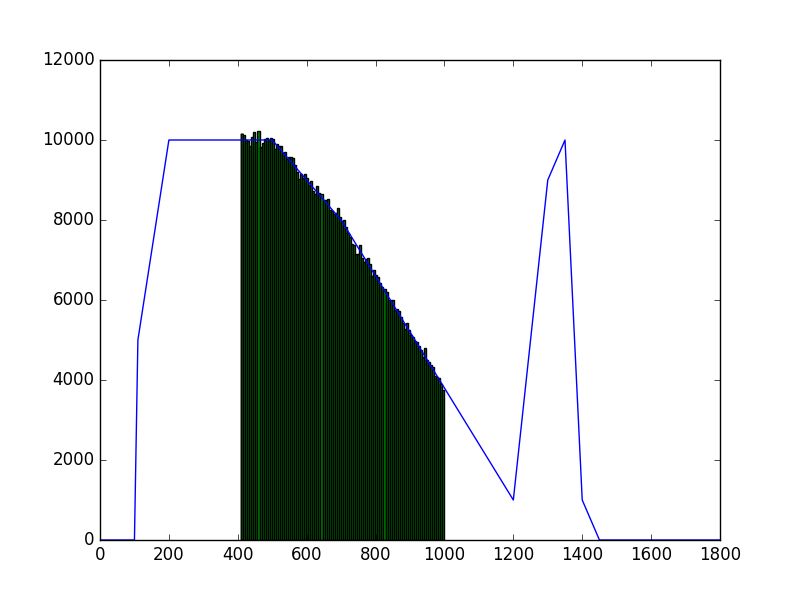
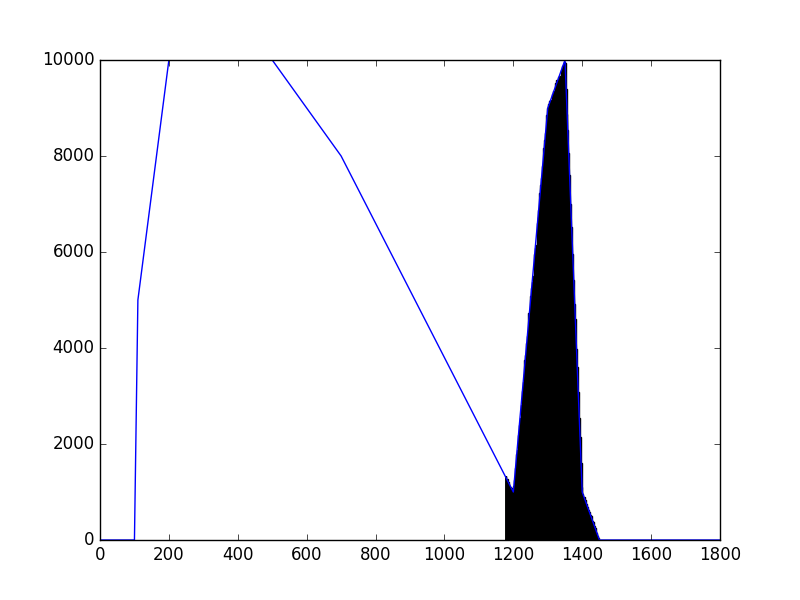
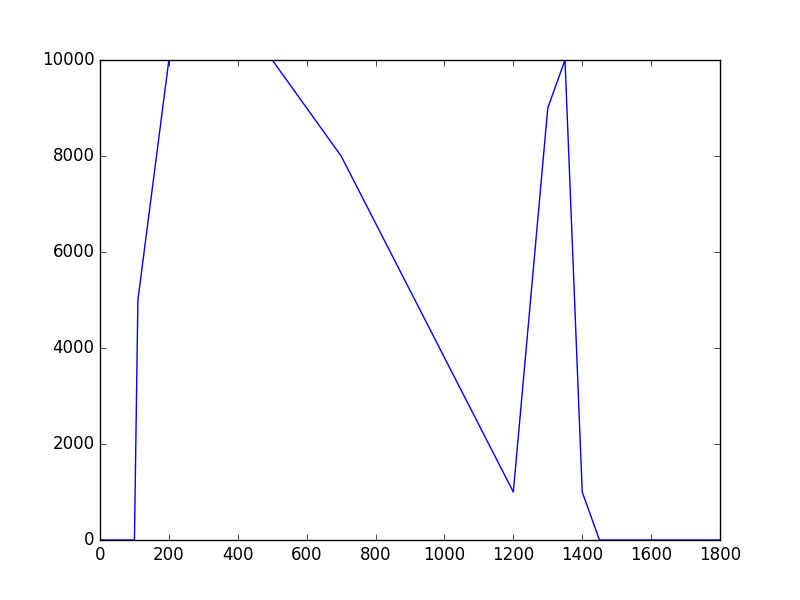
The class has been implemented, a dedicated unit test has been added and the mc() and eval() methods have been tested using a dedicated test script (attached). The visual output of the test script is shown below, indicating that the mc() method works properly for various time intervals.
 |
 |
 |
 |
 |
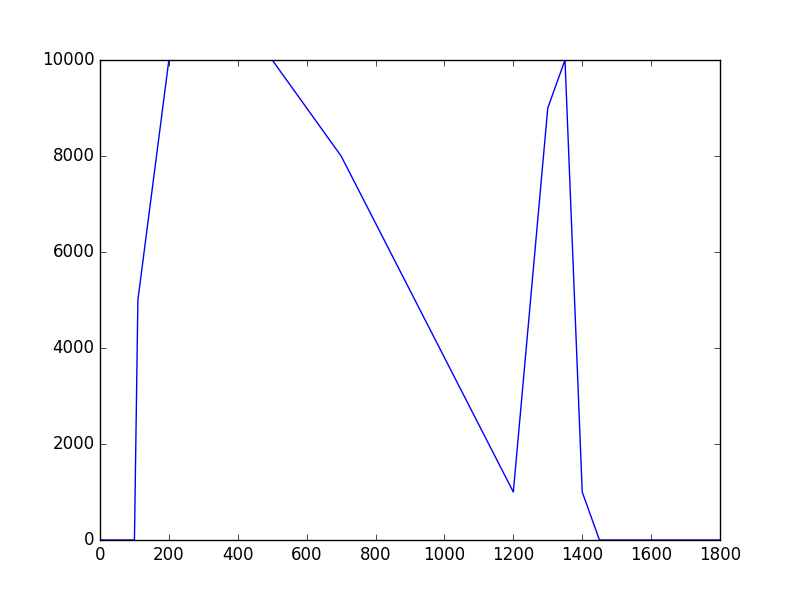 |
#2
 Updated by Knödlseder Jürgen about 8 years ago
Updated by Knödlseder Jürgen about 8 years ago
- File cslightcvr.png added
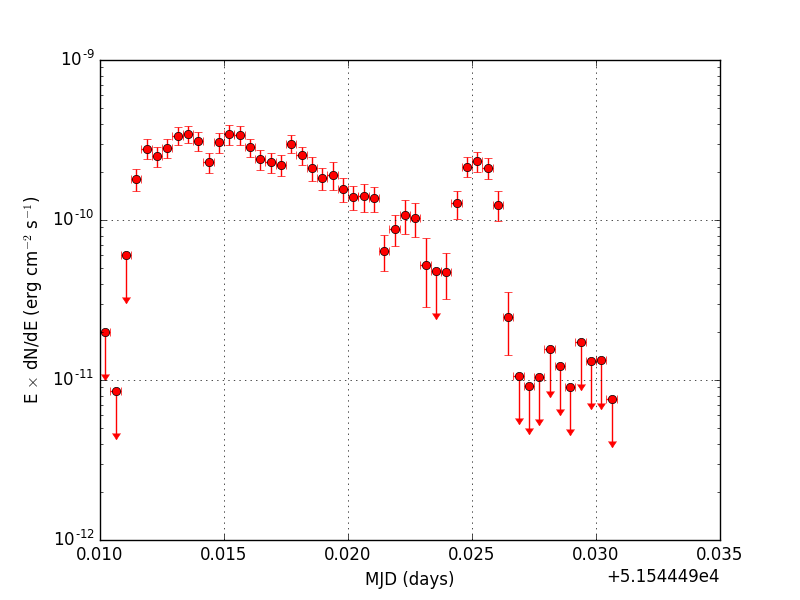
Below the light curve generated using the cslightcrv script from the event file simulated with ctobssim using a temporal file function. Note that the y-axis is logarithmic. The plot was generated using the show_lightcurve.py script.
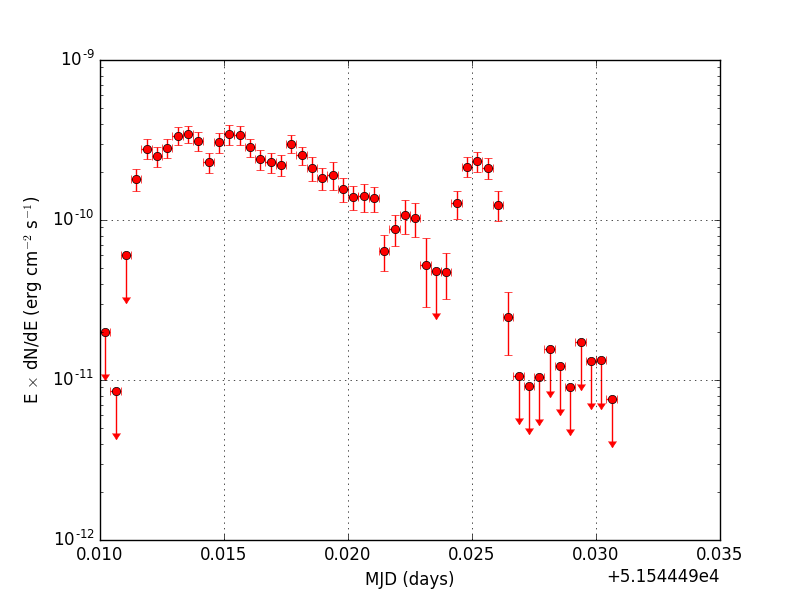 |
#3
 Updated by Knödlseder Jürgen about 8 years ago
Updated by Knödlseder Jürgen about 8 years ago
Finally, I also checked that the fitting of the temporal file function works. See the output below. I still need to generate pull distributions to see whether they are also okay.
2017-01-26T08:28:49: +=================================+ 2017-01-26T08:28:49: | Maximum likelihood optimisation | 2017-01-26T08:28:49: +=================================+ 2017-01-26T08:28:49: >Iteration 0: -logL=147599.390, Lambda=1.0e-03 2017-01-26T08:28:50: >Iteration 1: -logL=147595.361, Lambda=1.0e-03, delta=4.030, max(|grad|)=7.453374 [Index:3] 2017-01-26T08:28:50: >Iteration 2: -logL=147595.348, Lambda=1.0e-04, delta=0.012, max(|grad|)=0.022177 [Index:3] 2017-01-26T08:28:51: >Iteration 3: -logL=147595.348, Lambda=1.0e-05, delta=0.000, max(|grad|)=-0.000017 [Index:3] 2017-01-26T08:28:51: 2017-01-26T08:28:51: +=========================================+ 2017-01-26T08:28:51: | Maximum likelihood optimisation results | 2017-01-26T08:28:51: +=========================================+ 2017-01-26T08:28:51: === GOptimizerLM === 2017-01-26T08:28:51: Optimized function value ..: 147595.348 2017-01-26T08:28:51: Absolute precision ........: 0.005 2017-01-26T08:28:51: Acceptable value decrease .: 2 2017-01-26T08:28:51: Optimization status .......: converged 2017-01-26T08:28:51: Number of parameters ......: 10 2017-01-26T08:28:51: Number of free parameters .: 4 2017-01-26T08:28:51: Number of iterations ......: 3 2017-01-26T08:28:51: Lambda ....................: 1e-06 2017-01-26T08:28:51: Maximum log likelihood ....: -147595.348 2017-01-26T08:28:51: Observed events (Nobs) ...: 21022.000 2017-01-26T08:28:51: Predicted events (Npred) ..: 21022.000 (Nobs - Npred = 1.61784555530176e-07) 2017-01-26T08:28:51: === GModels === 2017-01-26T08:28:51: Number of models ..........: 2 2017-01-26T08:28:51: Number of parameters ......: 10 2017-01-26T08:28:51: === GModelSky === 2017-01-26T08:28:51: Name ......................: Crab 2017-01-26T08:28:51: Instruments ...............: all 2017-01-26T08:28:51: Instrument scale factors ..: unity 2017-01-26T08:28:51: Observation identifiers ...: all 2017-01-26T08:28:51: Model type ................: PointSource 2017-01-26T08:28:51: Model components ..........: "PointSource" * "PowerLaw" * "FileFunction" 2017-01-26T08:28:51: Number of parameters ......: 6 2017-01-26T08:28:51: Number of spatial par's ...: 2 2017-01-26T08:28:51: RA .......................: 83.6331 [-360,360] deg (fixed,scale=1) 2017-01-26T08:28:51: DEC ......................: 22.0145 [-90,90] deg (fixed,scale=1) 2017-01-26T08:28:51: Number of spectral par's ..: 3 2017-01-26T08:28:51: Prefactor ................: 5.64109934986393e-16 +/- 1.44694059813623e-17 [1e-23,1e-13] ph/cm2/s/MeV (free,scale=1e-16,gradient) 2017-01-26T08:28:51: Index ....................: -2.42121810308988 +/- 0.020974164555023 [-0,-5] (free,scale=-1,gradient) 2017-01-26T08:28:51: PivotEnergy ..............: 300000 [10000,1000000000] MeV (fixed,scale=1000000,gradient) 2017-01-26T08:28:51: Number of temporal par's ..: 1 2017-01-26T08:28:51: Normalization ............: 1 [0,1000] (relative value) (fixed,scale=1,gradient) 2017-01-26T08:28:51: === GCTAModelIrfBackground === 2017-01-26T08:28:51: Name ......................: CTABackgroundModel 2017-01-26T08:28:51: Instruments ...............: CTA 2017-01-26T08:28:51: Instrument scale factors ..: unity 2017-01-26T08:28:51: Observation identifiers ...: all 2017-01-26T08:28:51: Model type ................: "PowerLaw" * "Constant" 2017-01-26T08:28:51: Number of parameters ......: 4 2017-01-26T08:28:51: Number of spectral par's ..: 3 2017-01-26T08:28:51: Prefactor ................: 0.996001391554557 +/- 0.0118927869244997 [0.001,1000] ph/cm2/s/MeV (free,scale=1,gradient) 2017-01-26T08:28:51: Index ....................: -0.00066143101849483 +/- 0.0073650960488301 [-5,5] (free,scale=1,gradient) 2017-01-26T08:28:51: PivotEnergy ..............: 1000000 [10000,1000000000] MeV (fixed,scale=1000000,gradient) 2017-01-26T08:28:51: Number of temporal par's ..: 1 2017-01-26T08:28:51: Normalization ............: 1 (relative value) (fixed,scale=1,gradient)
#4
 Updated by Knödlseder Jürgen about 8 years ago
Updated by Knödlseder Jürgen about 8 years ago
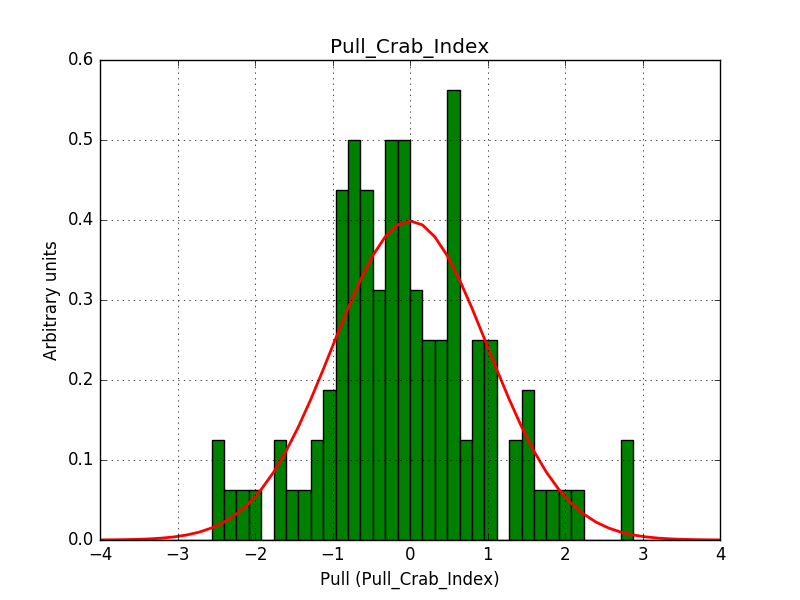
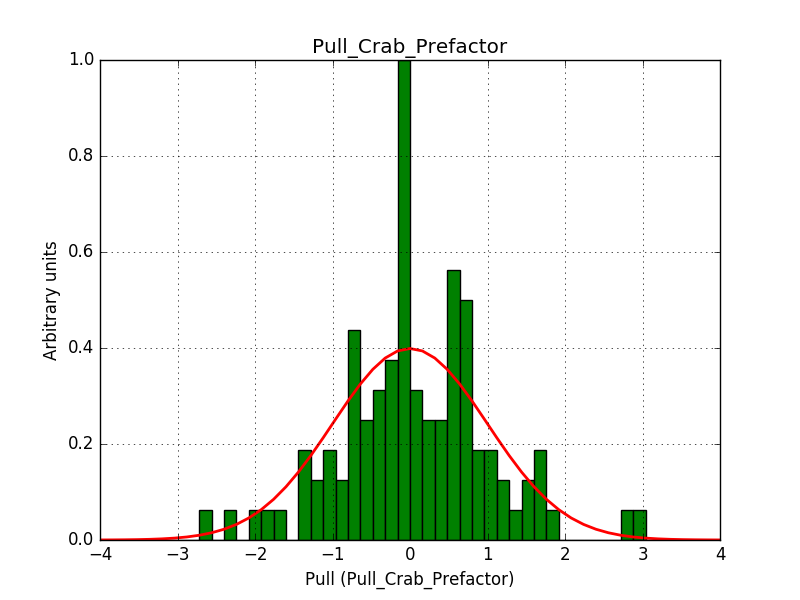
- File pull_prefactor.png added
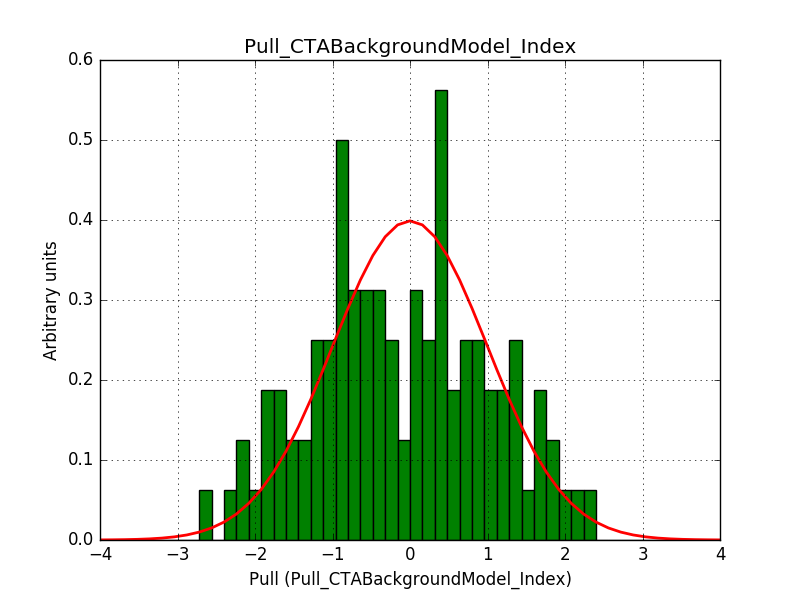
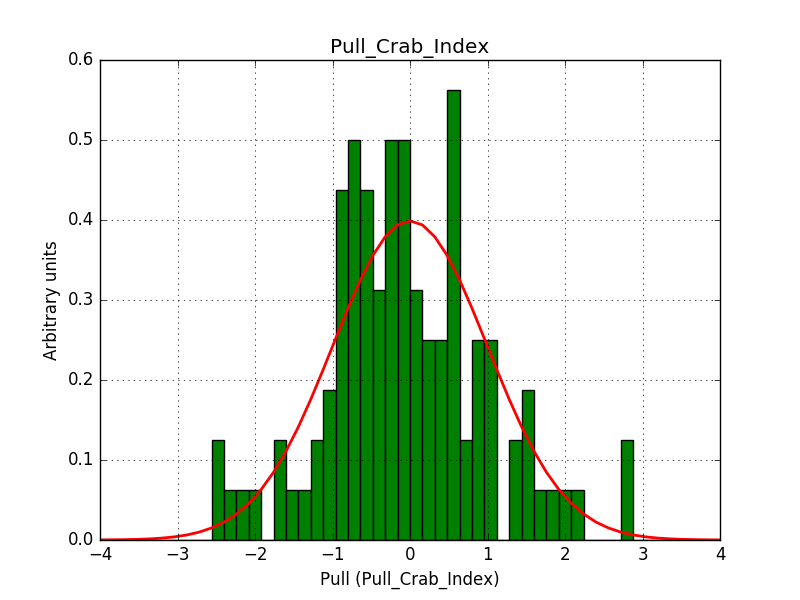
- File pull_index.png added
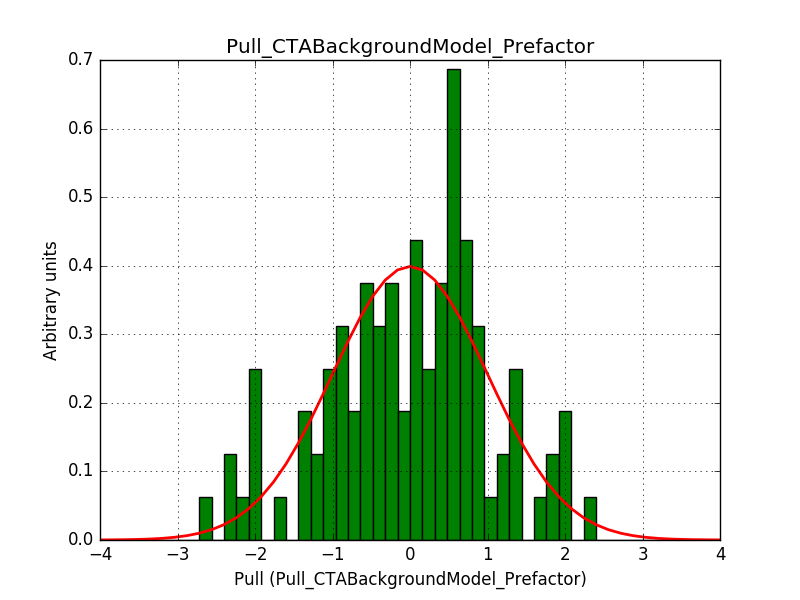
- File pull_bgd_prefactor.png added
- File pull_bgd_index.png added
- Status changed from New to Closed
- % Done changed from 90 to 100
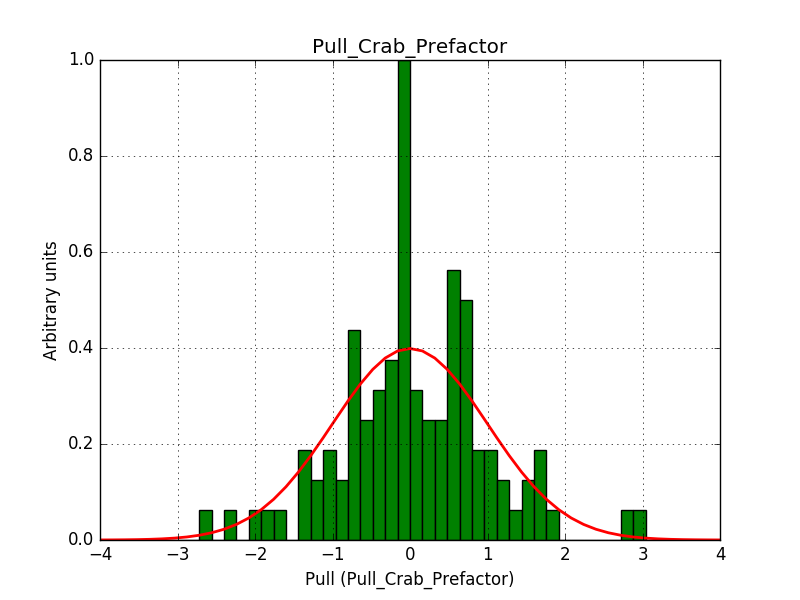
Also the pull distributions look good. Consider that this model is now validated. Close the issue.
 |
 |
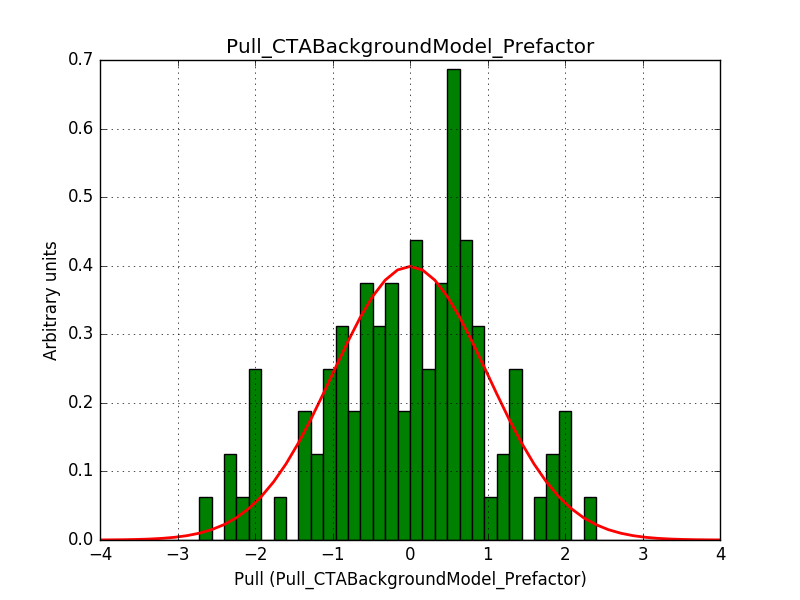 |
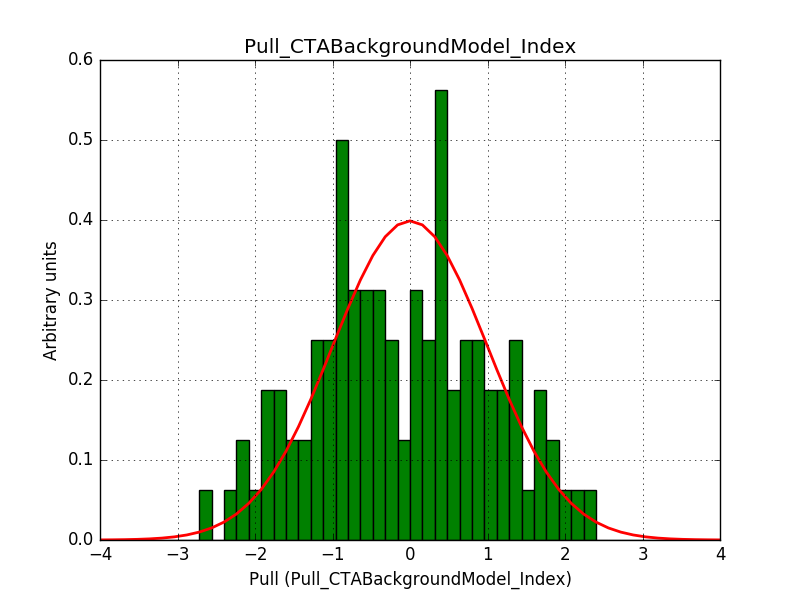 |