Action #3501
Implement vectorised response computation for COMPTEL
| Status: | Closed | Start date: | 01/15/2021 | |
|---|---|---|---|---|
| Priority: | Normal | Due date: | ||
| Assigned To: | % Done: | 100% | ||
| Category: | - | |||
| Target version: | 2.0.0 | |||
| Duration: |
Recurrence
No recurrence.
History
#1
 Updated by Knödlseder Jürgen about 4 years ago
Updated by Knödlseder Jürgen about 4 years ago
- Status changed from New to In Progress
- Assigned To set to Knödlseder Jürgen
- % Done changed from 0 to 50
I implemented a vectorised response computation in GCOMResponse::irf_ptsrc() making use of the characteristics of the COMPTEL data space. Specifically, phigeo is now only computed once for the Chi/Psi pixel.
Initial testing using a five energy-band Crab fit reduced the ctlike execution time from 23.23 seconds to 12.48 seconds, hence leading to a speed-up of a factor of 2. More testing is needed to verify for example that the code also works for TS map computation. Also, a valgrind analysis may be done to further speed-up the analysis.
#2
 Updated by Knödlseder Jürgen about 4 years ago
Updated by Knödlseder Jürgen about 4 years ago
- % Done changed from 50 to 60
There is basically no speed-up in the spectral analysis, which is normal since the spatial parameters are fixed in the spectral analysis. Consequently, the cached values will be used.
#3
 Updated by Knödlseder Jürgen about 4 years ago
Updated by Knödlseder Jürgen about 4 years ago
- % Done changed from 60 to 70
The same holds for the TS map computation. No speed-up because the spatial parameters are fixed.
#4
 Updated by Knödlseder Jürgen about 4 years ago
Updated by Knödlseder Jürgen about 4 years ago
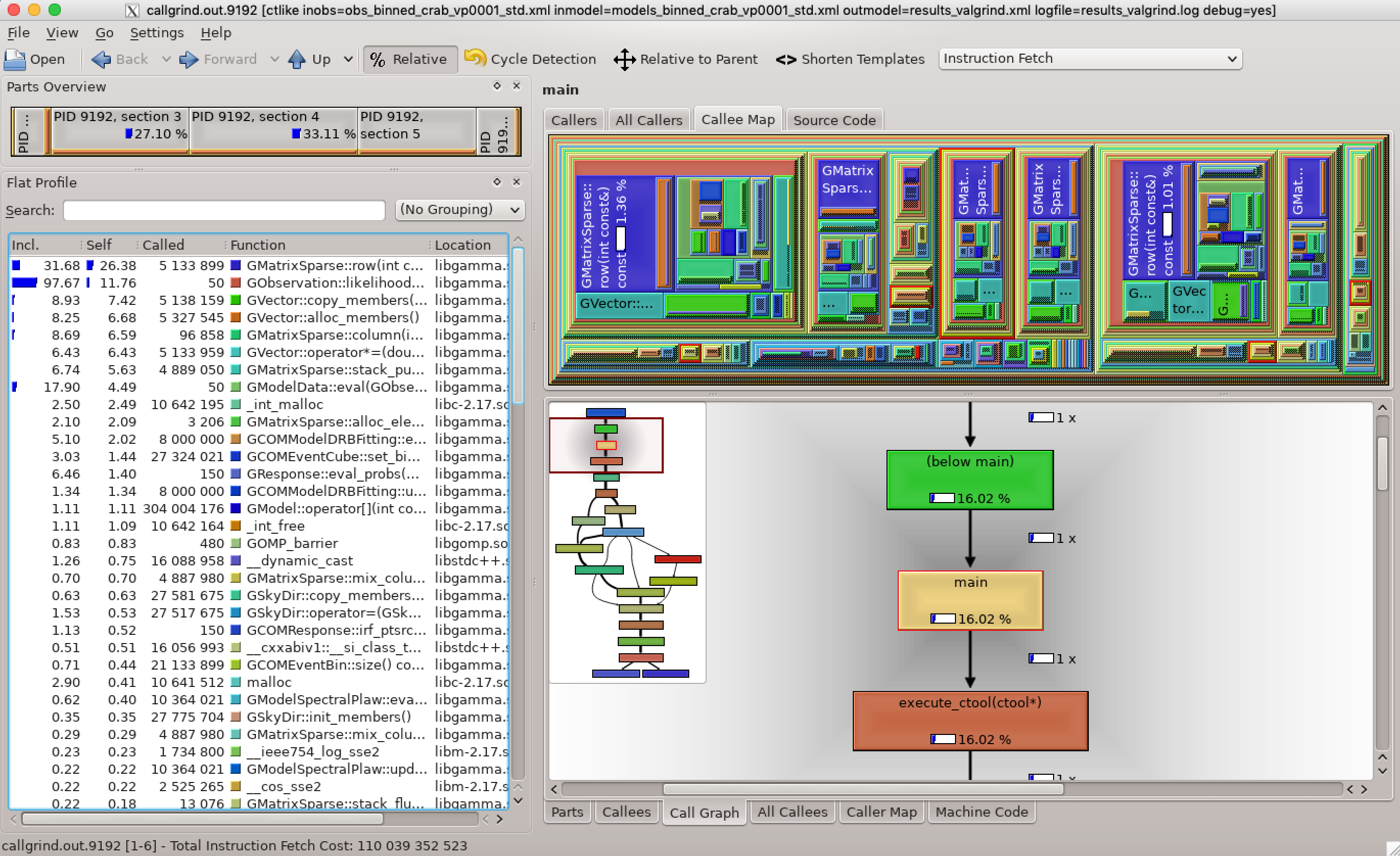
- File kcachegrind.png added
Here is a valgrind analysis of the 5 standard band ctlike run on viewing period 1. Obviously a significant fraction of time is spent in the matrix and vector handling.
Specifically, the GMatrixSparse::row method is called many times in GObservation::likelihood_poisson_binned which uses quite some computing time. The reason for this is that the matrices are stored column-wise, hence row access needs quite some computations. It should be checked whether a transposition of the matrix and a column access residuals in actually faster code.
#5
 Updated by Knödlseder Jürgen about 4 years ago
Updated by Knödlseder Jürgen about 4 years ago
- % Done changed from 70 to 80
Transposing the matrix in GObservation::likelihood_poisson_binned and accessing it through the GMatrixSparse::column method leads to faster code, using 10.21 instead of 12.50 seconds for the 5-band ctlike fit of viewing period 1. For the TS map, the computation needed 3671.03 seconds instead of 5017.22 seconds. Overall, a speed-up of about 20-25% is achieved.
I also checked the speed-up for fitting the Crab to simulated CTA data (fixed position, only prefactor and spectral index are free). Below the computation times and speed-up for the various analysis methods:
| Method | Original | Transposed | Speed-up |
| Unbinned | 7.95 s | 7.34 s | 8% |
| Binned | 5.97 s | 5.62 s | 6% |
| Stacked | 3.68 s | 3.31 s | 10% |
In all cases the code is faster. I will therefore implement the matrix transformation.
#6
 Updated by Knödlseder Jürgen about 4 years ago
Updated by Knödlseder Jürgen about 4 years ago
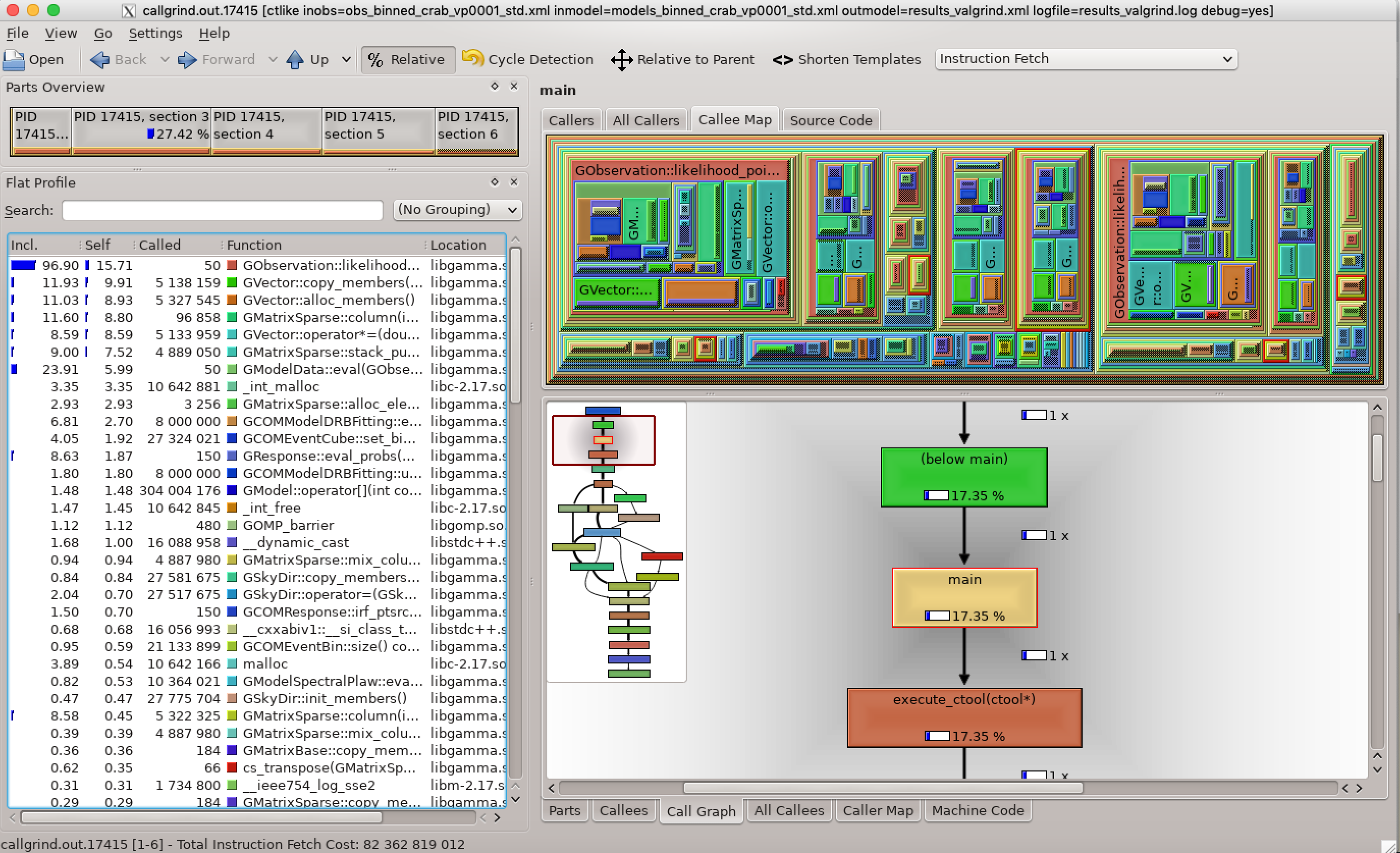
- File kcachegrind-transpose.png added
Below the valgrind analysis for the transposed matrix. As expected, matrix access is no longer a bottleneck (the GMatrixSparse::column method uses less than 1% of the time).
#7
 Updated by Knödlseder Jürgen about 4 years ago
Updated by Knödlseder Jürgen about 4 years ago
- Status changed from In Progress to Pull request
- % Done changed from 80 to 90
#8
 Updated by Knödlseder Jürgen about 4 years ago
Updated by Knödlseder Jürgen about 4 years ago
- Status changed from Pull request to Closed
- % Done changed from 90 to 100
Merged into devel.