Wiki » Development Notes »
Updated about 12 years ago by  Knödlseder Jürgen
Knödlseder Jürgen
GModelSpectralNodes¶
Validation¶
The model has been validation by generating pull distributions using cspull. The validation has been done using ctools-00-06-00 and GammaLib-00-07-00.
A power law following the HESS Crab spectrum was used to generate a 10 node spectral node model. The energy values of the 10 nodes are:
0.5, 0.834, 1.391, 2.321, 3.871, 6.458, 10.772, 17.969, 29.974, and 50 TeV.
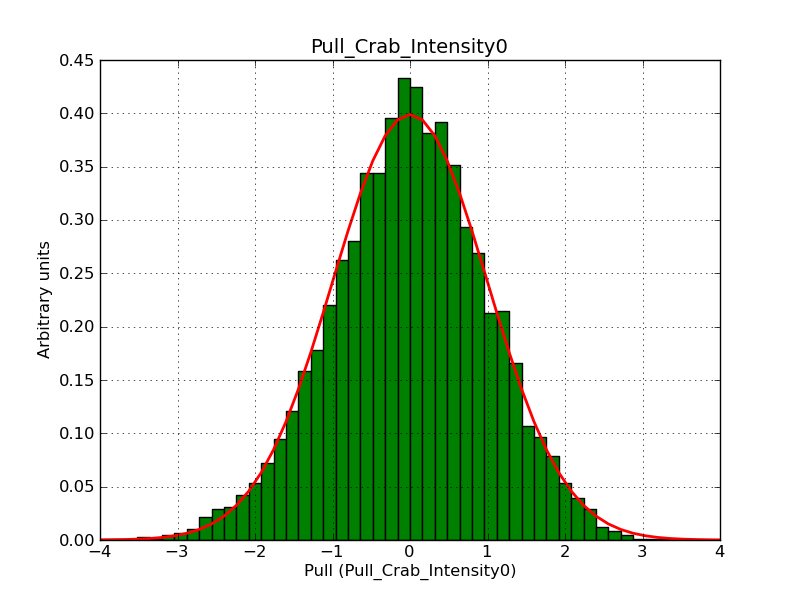
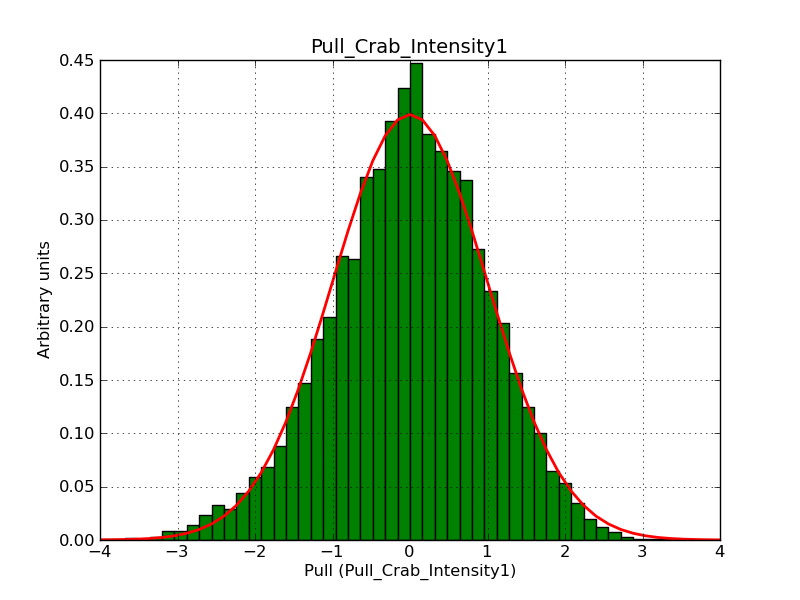
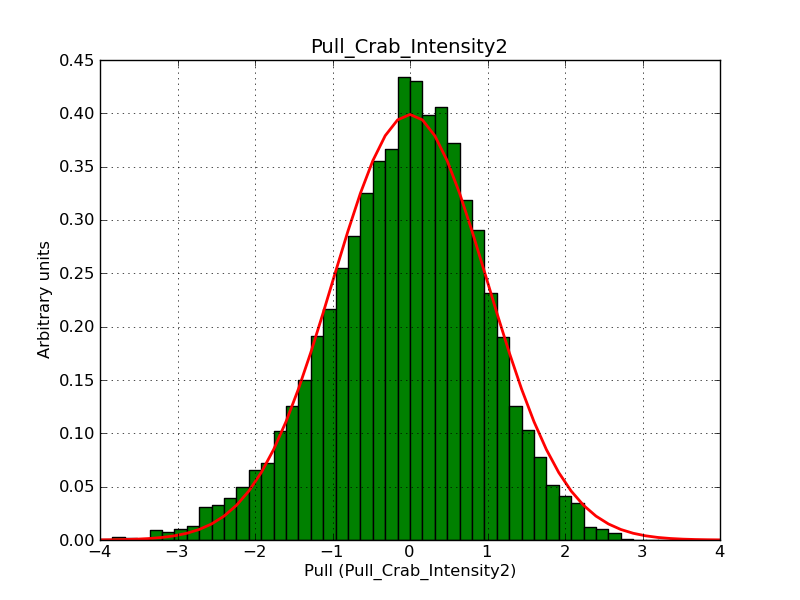
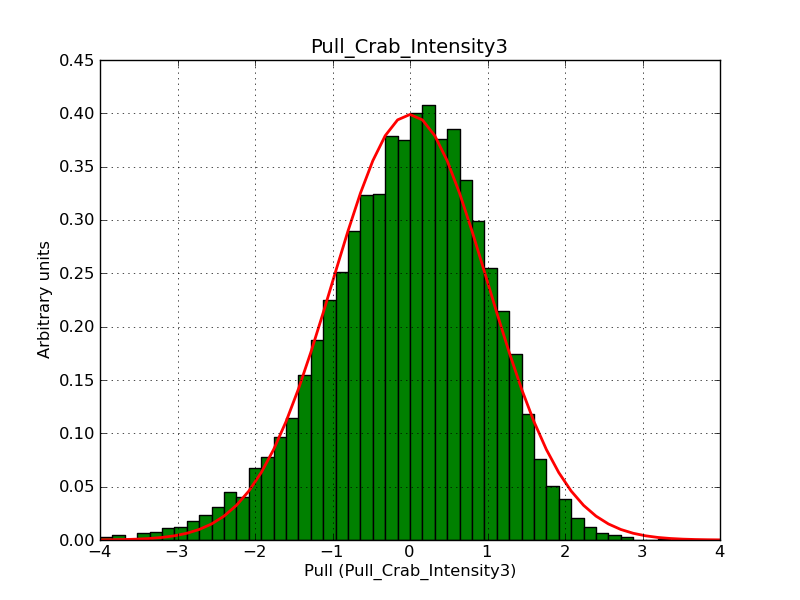
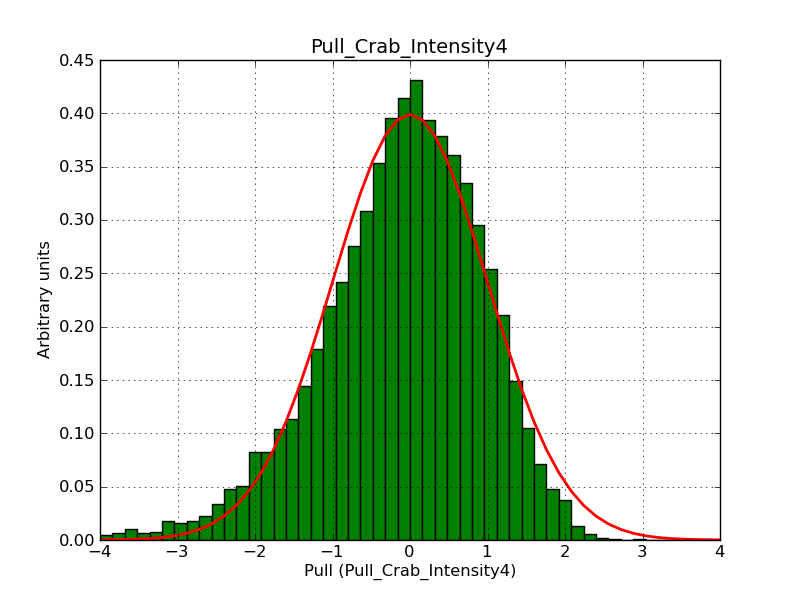
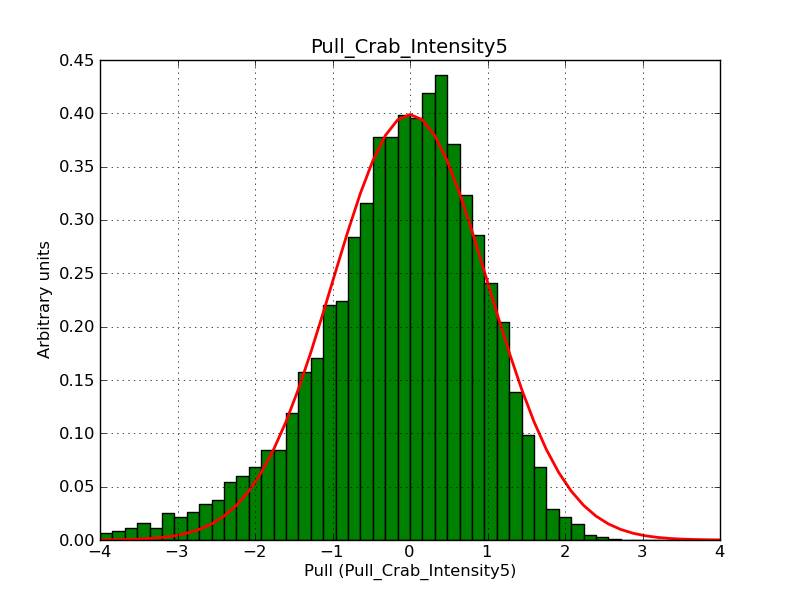
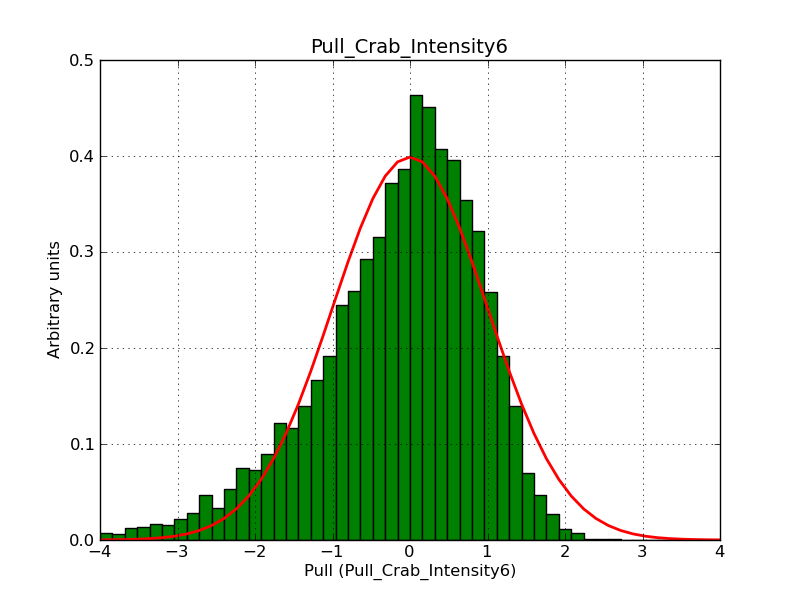
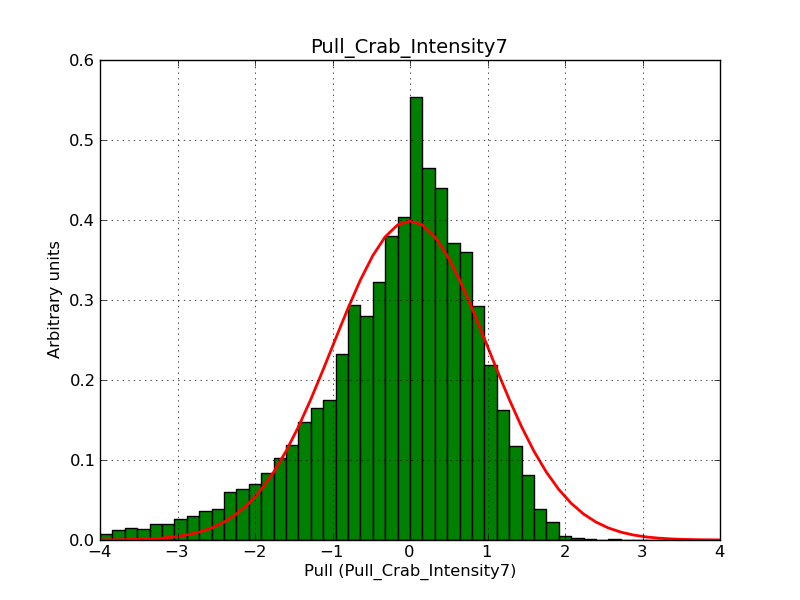
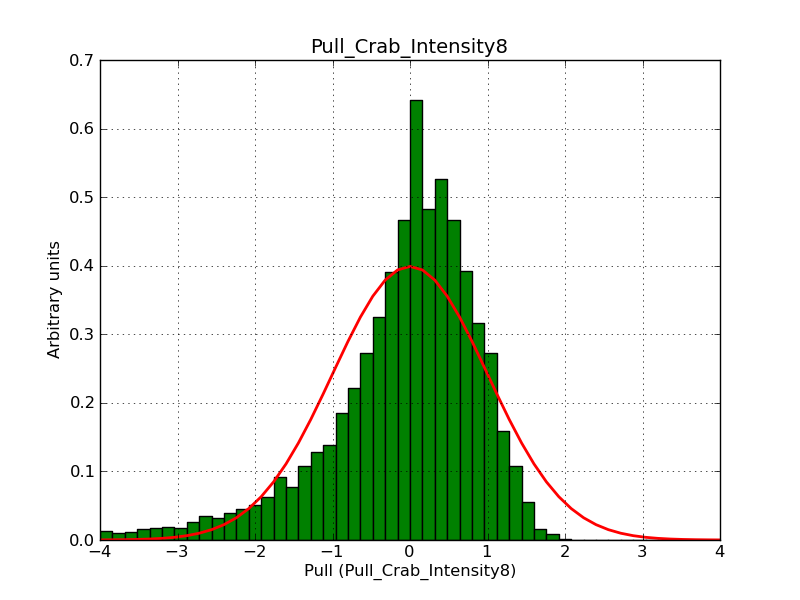
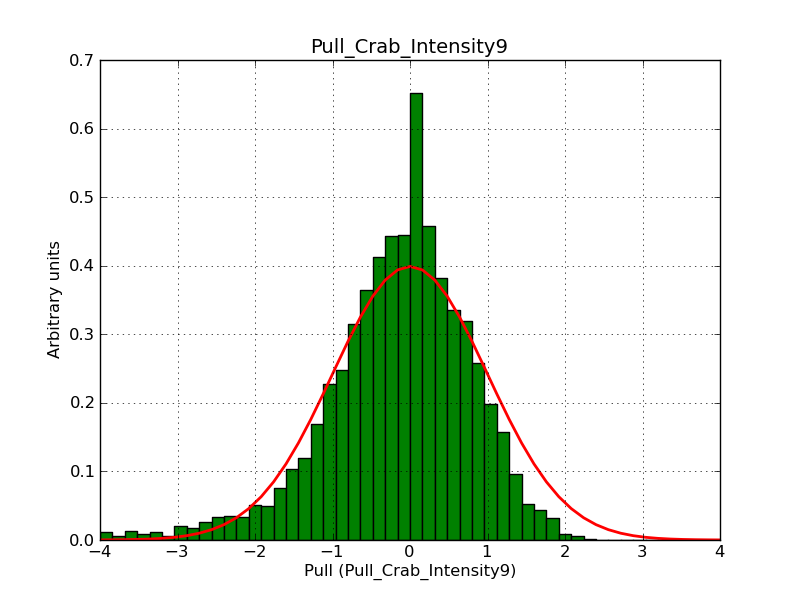
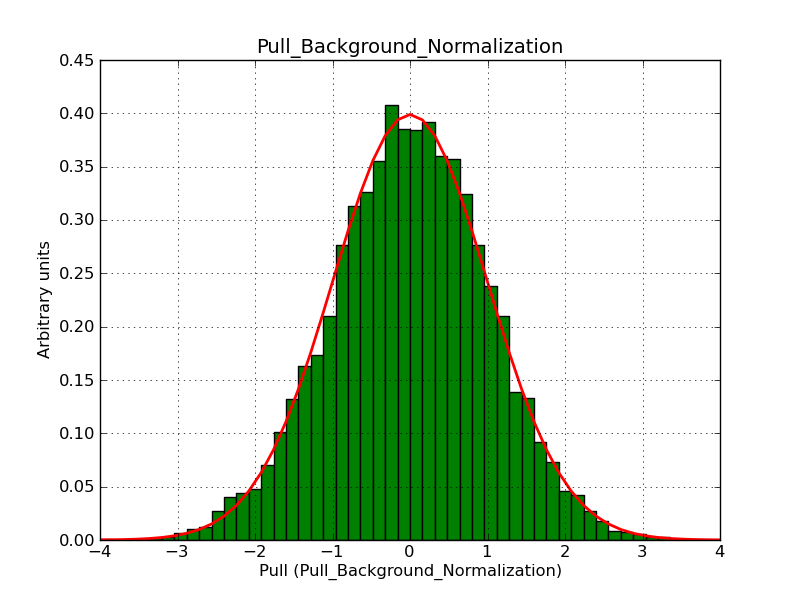
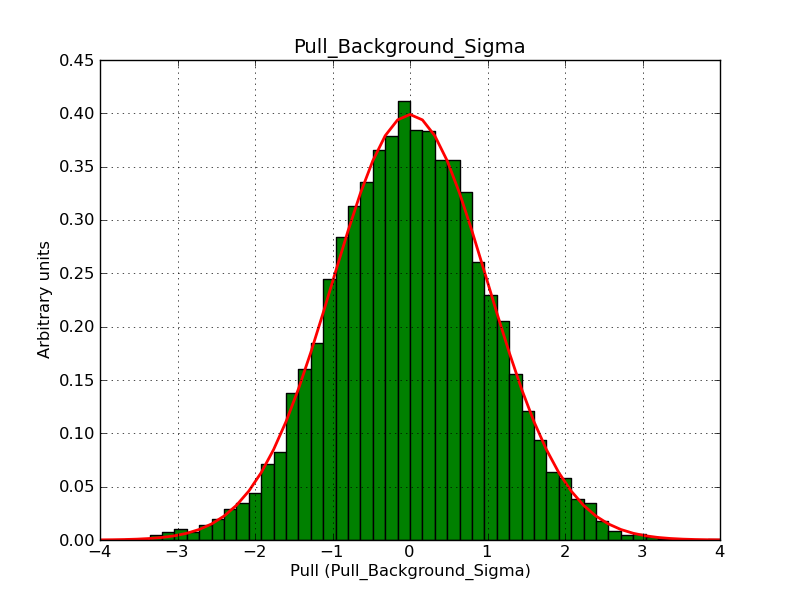
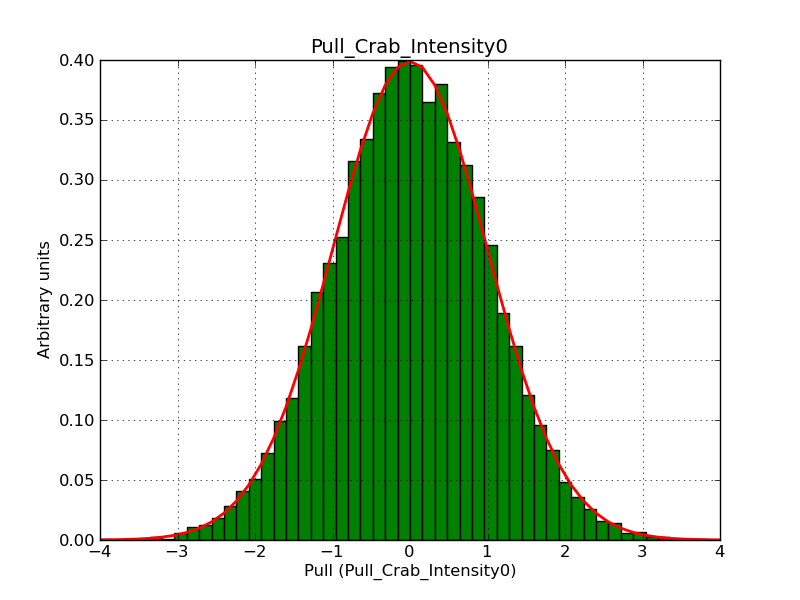
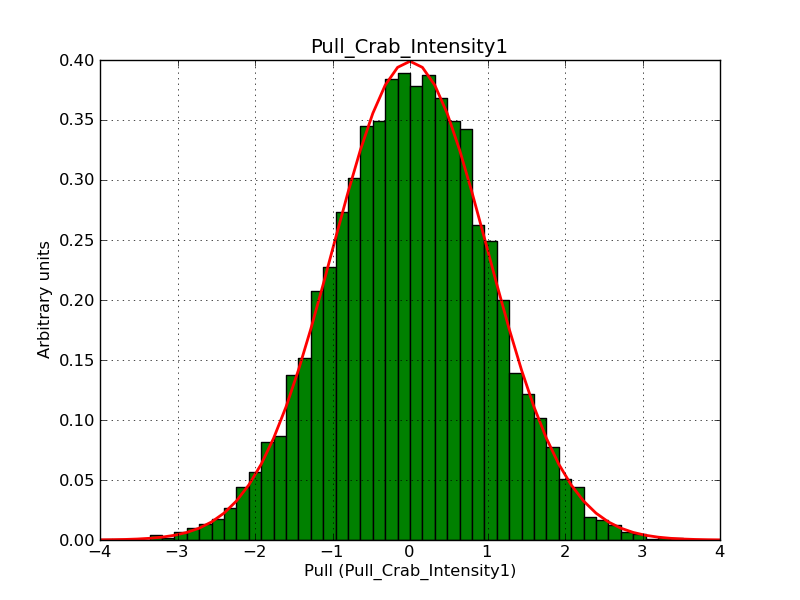
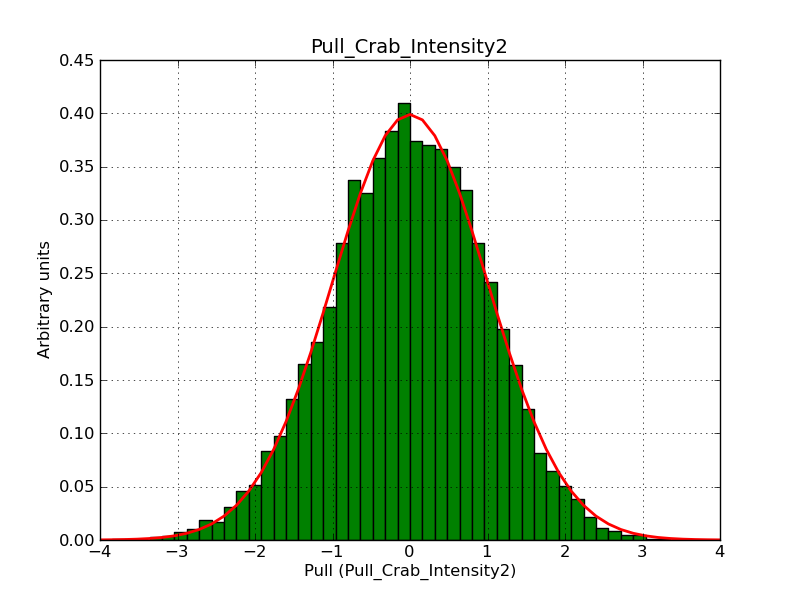
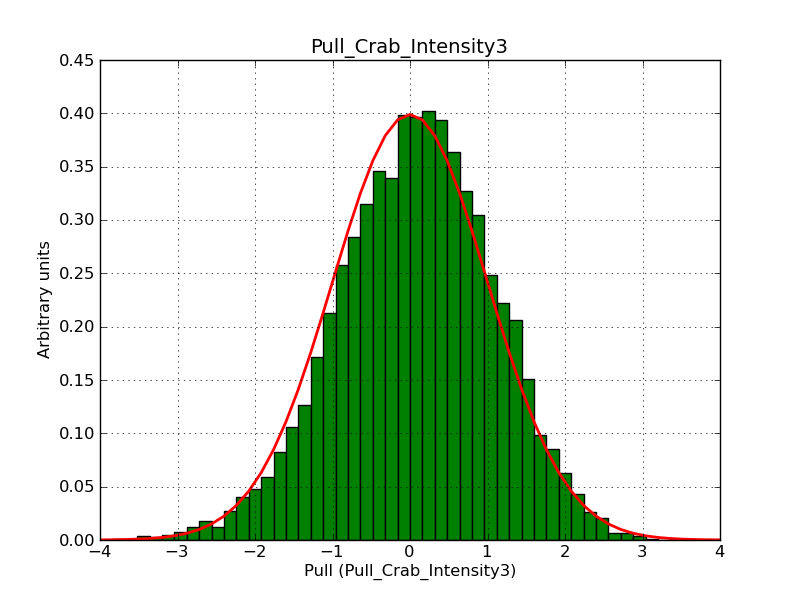
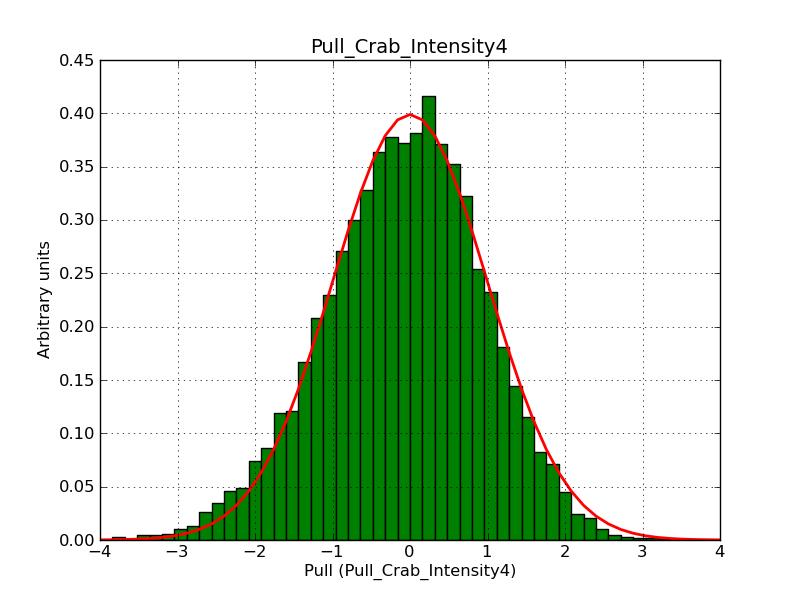
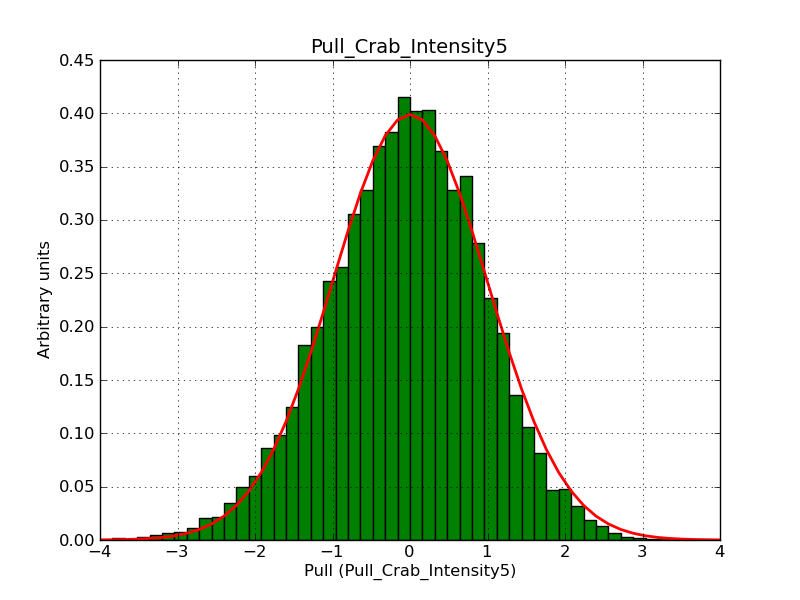
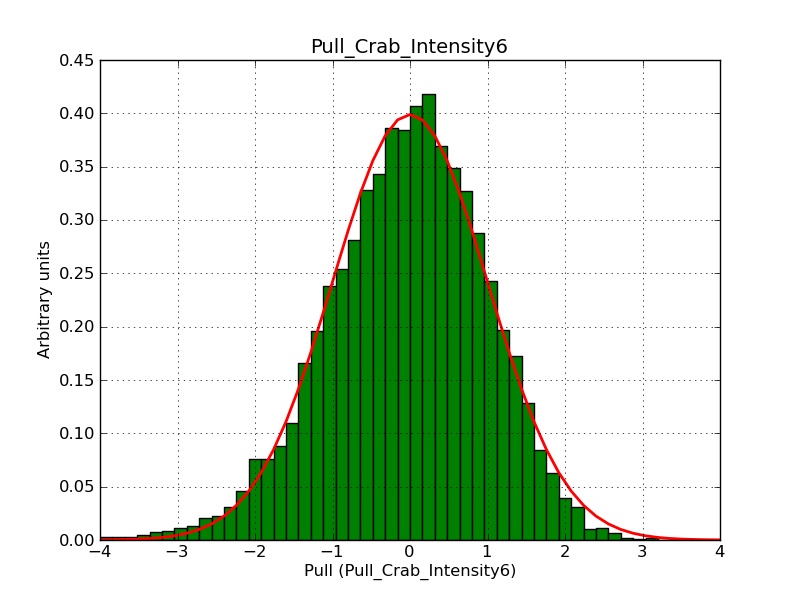
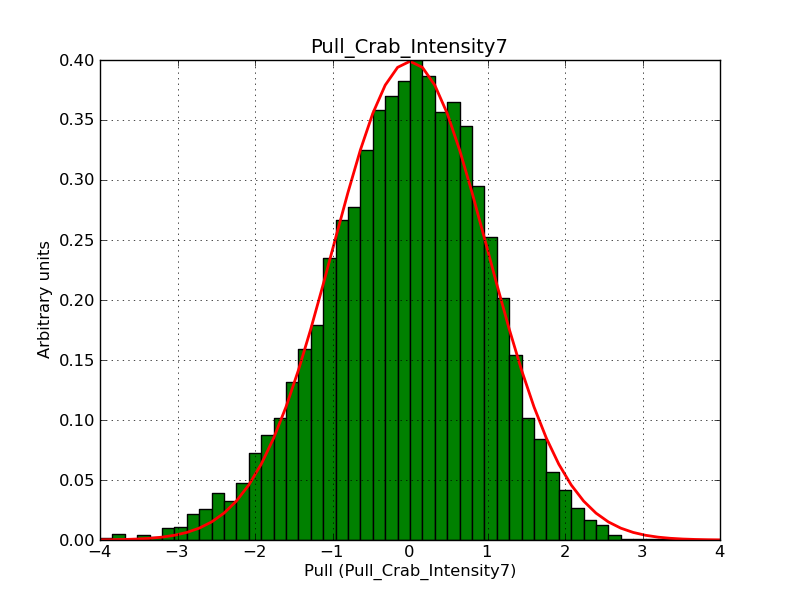
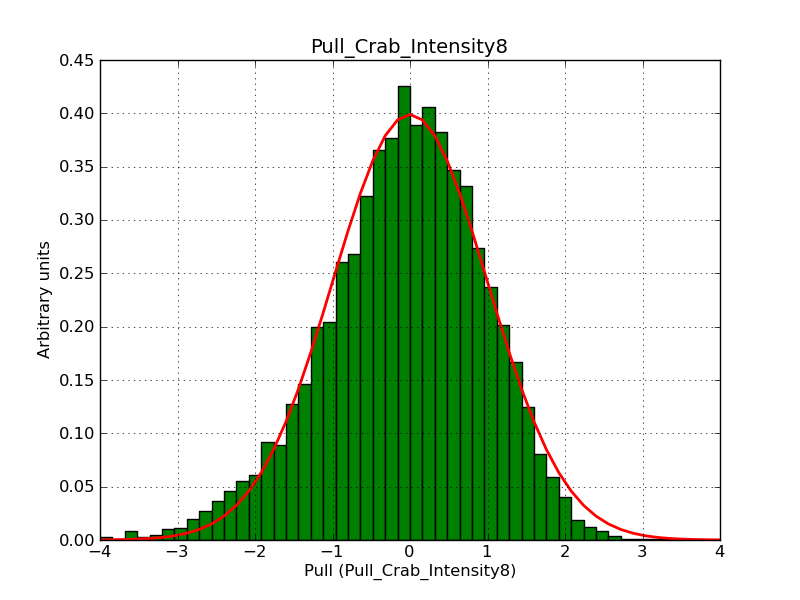
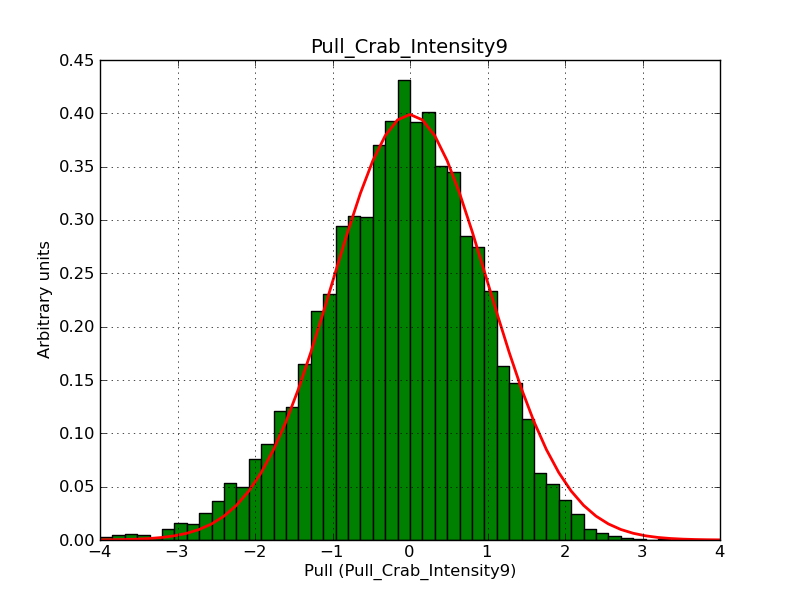
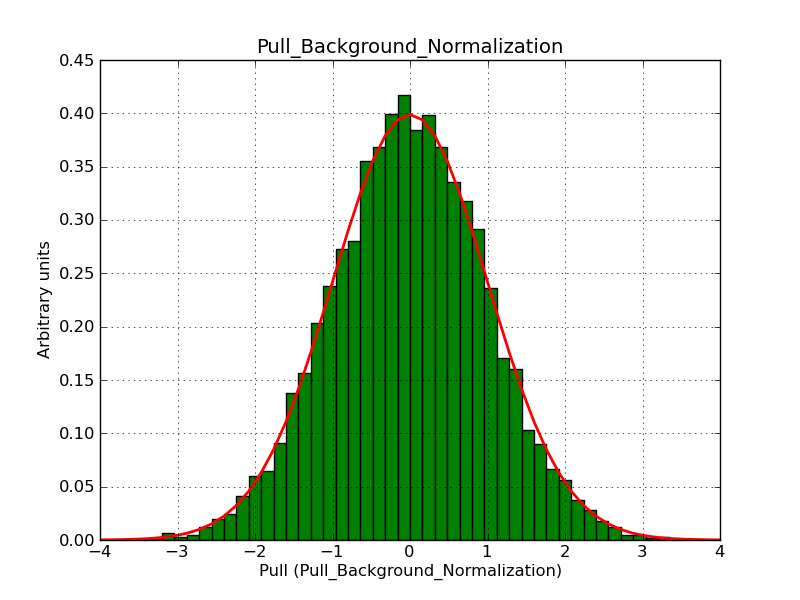
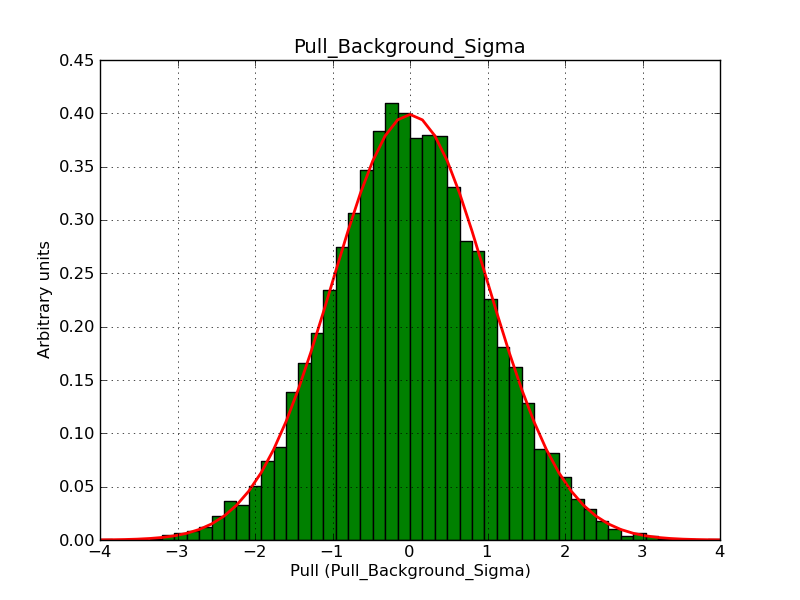
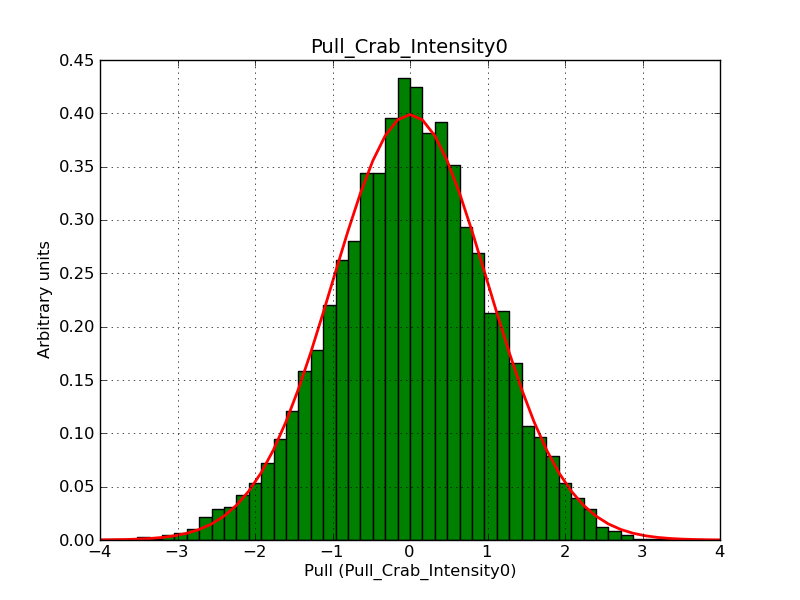
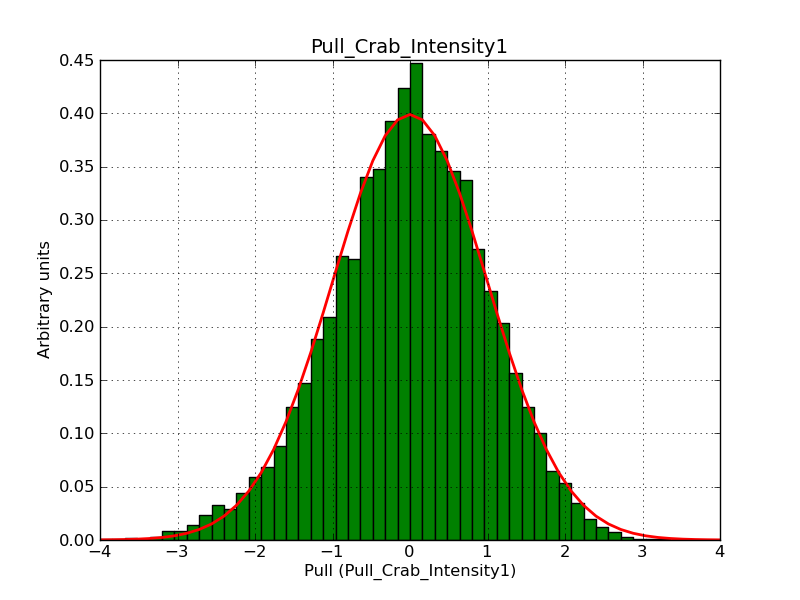
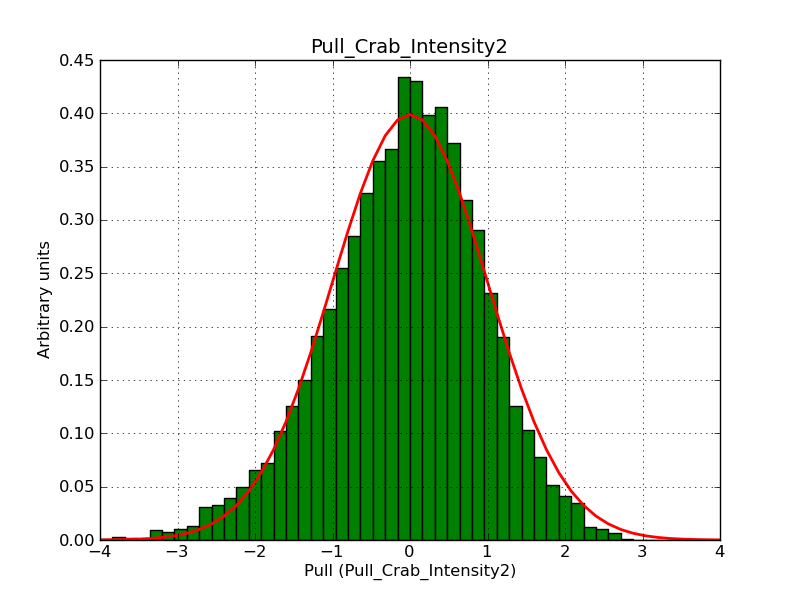
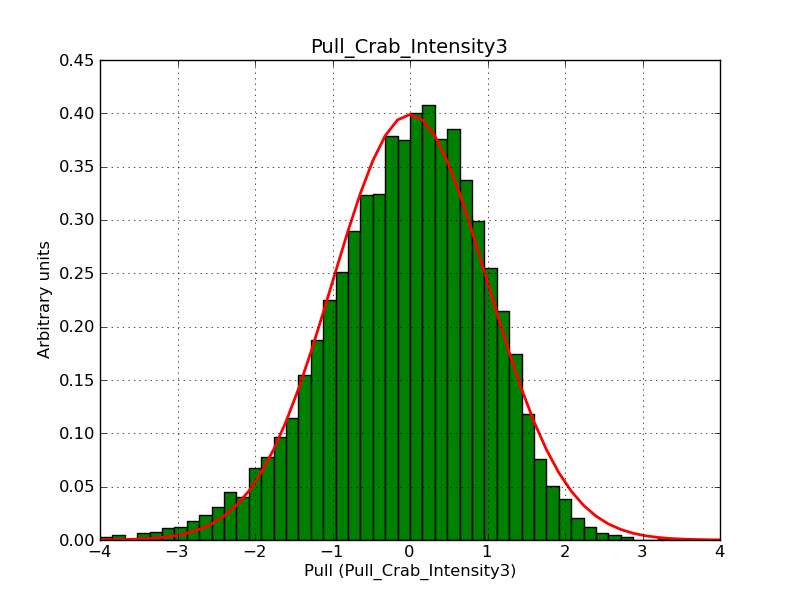
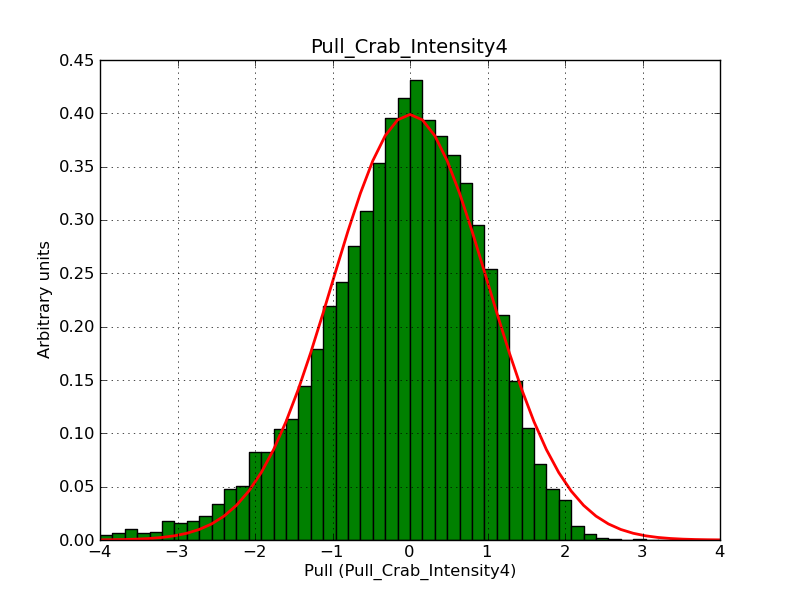
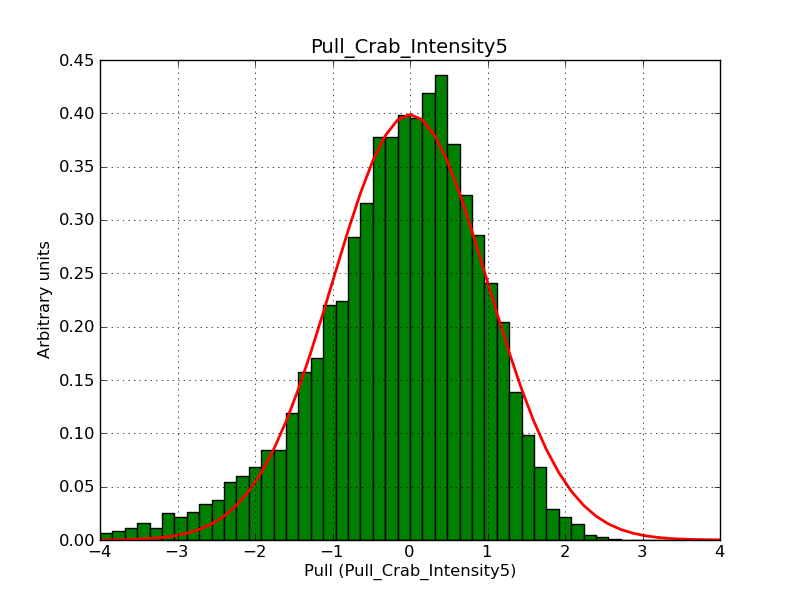
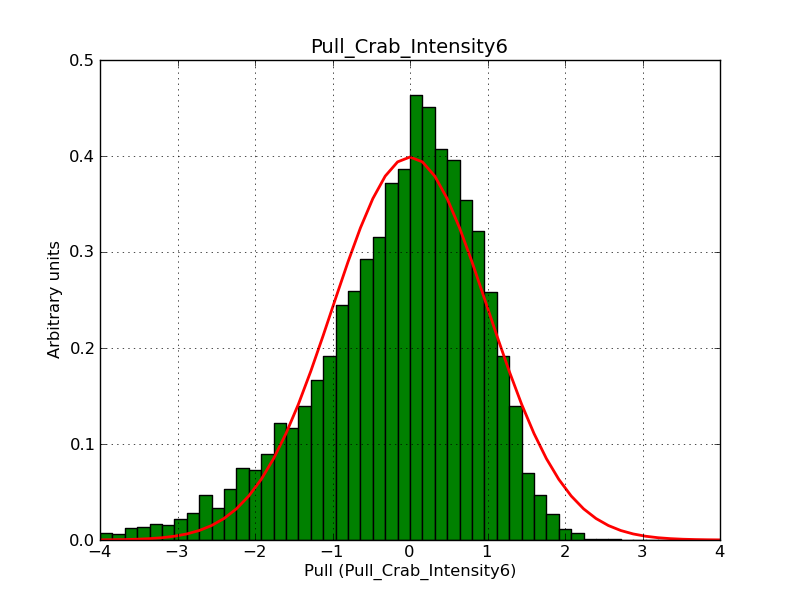
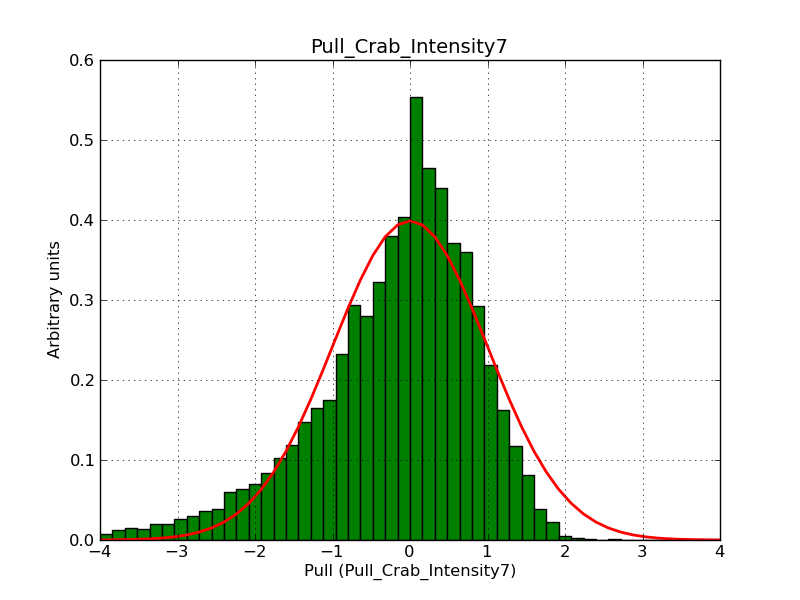
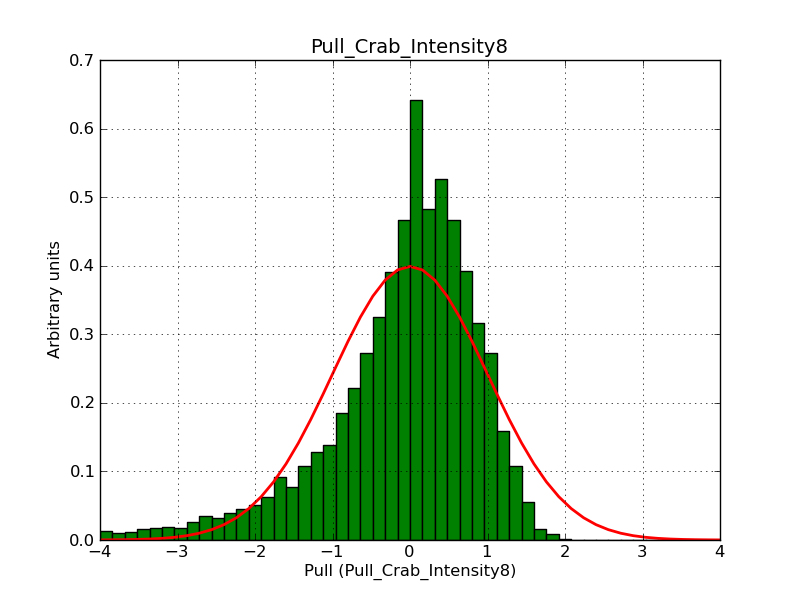
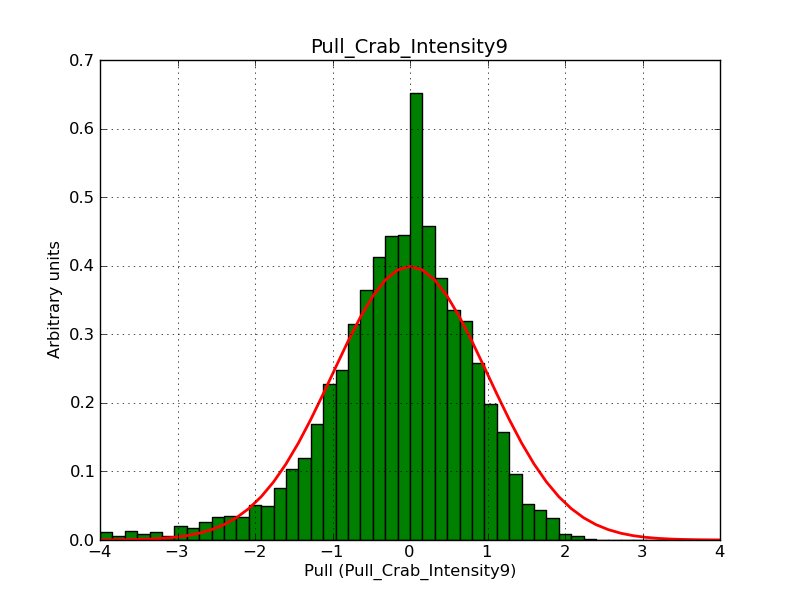
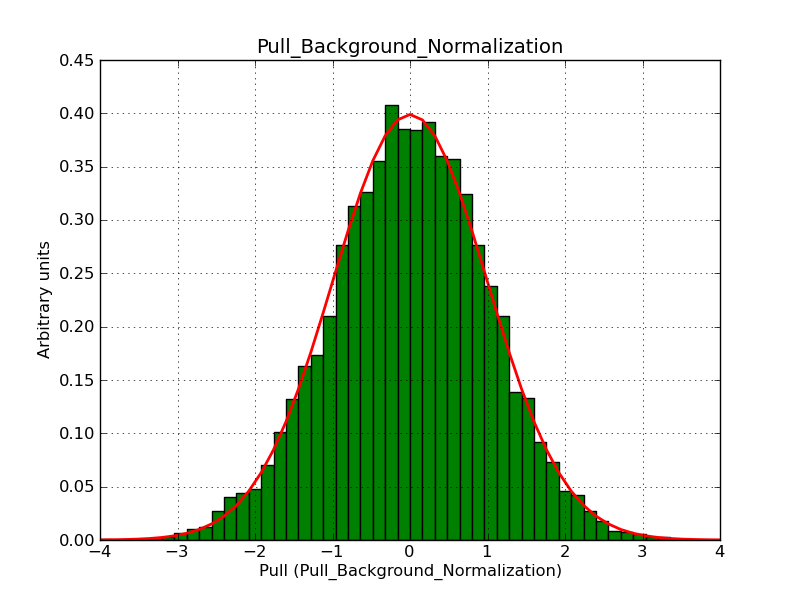
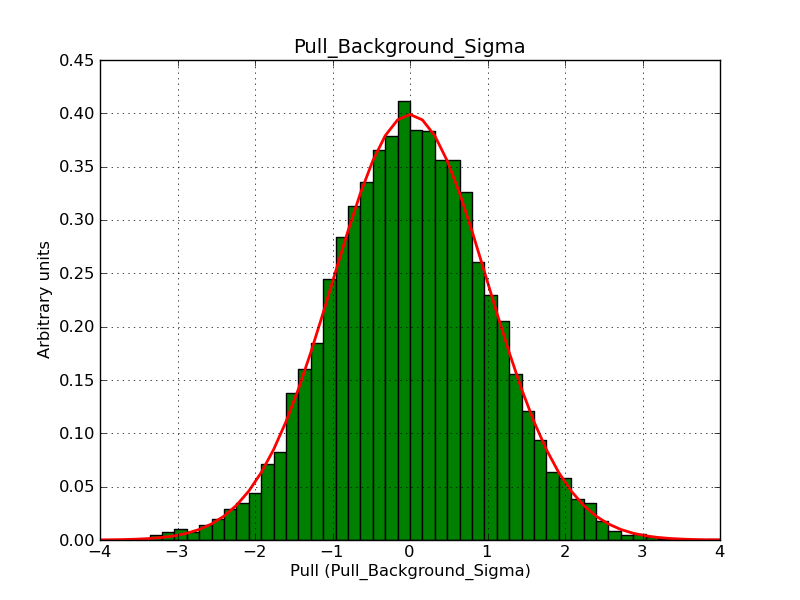
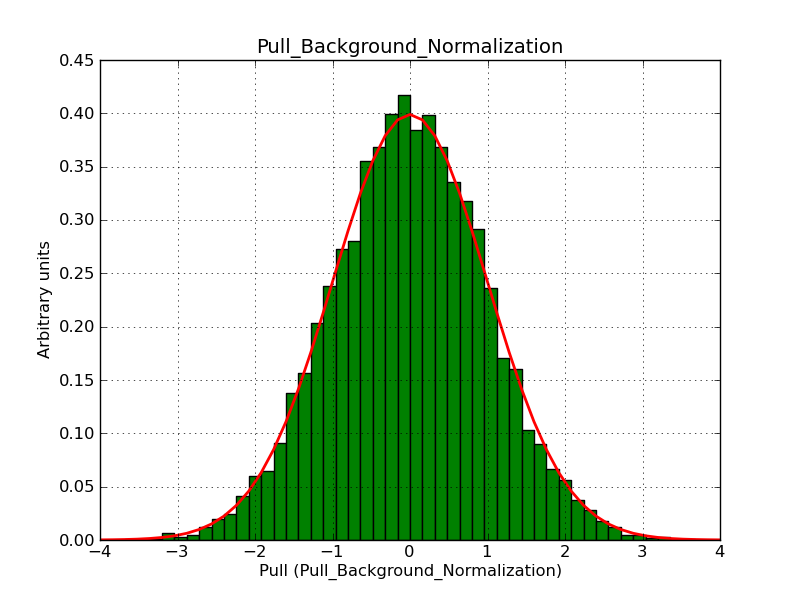
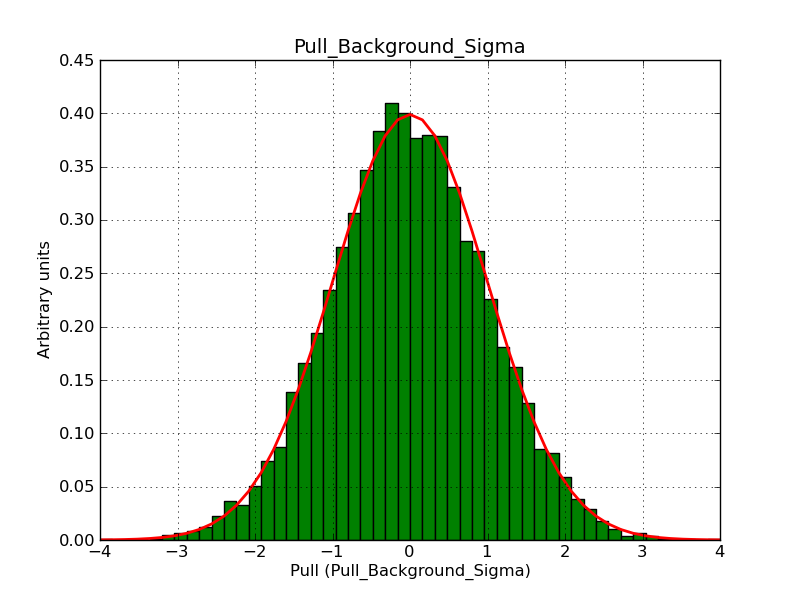
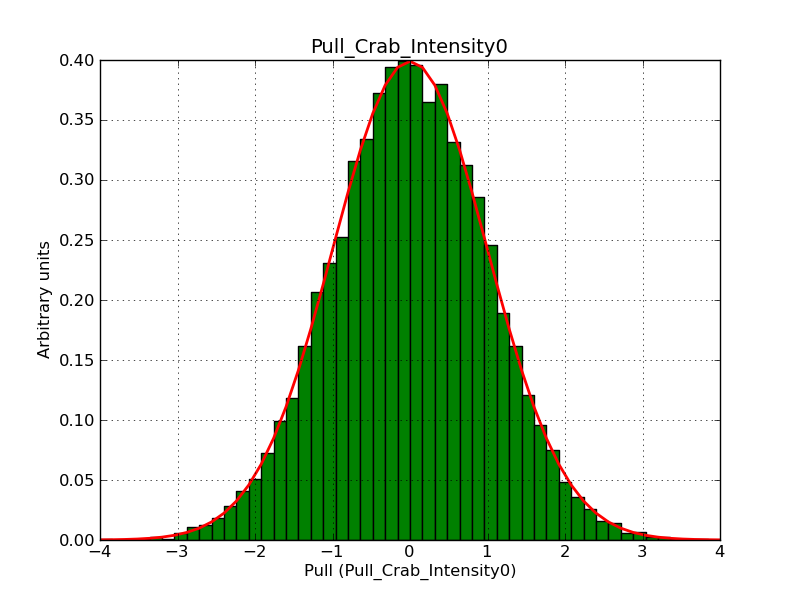
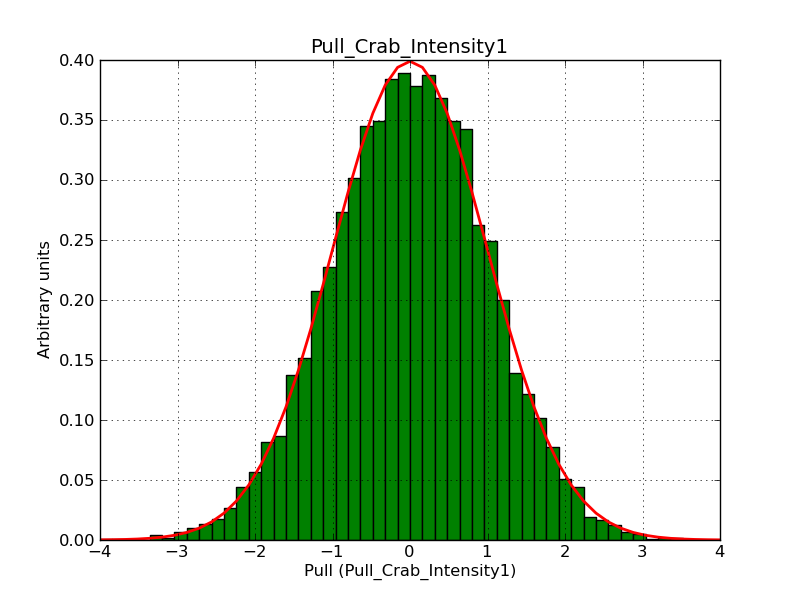
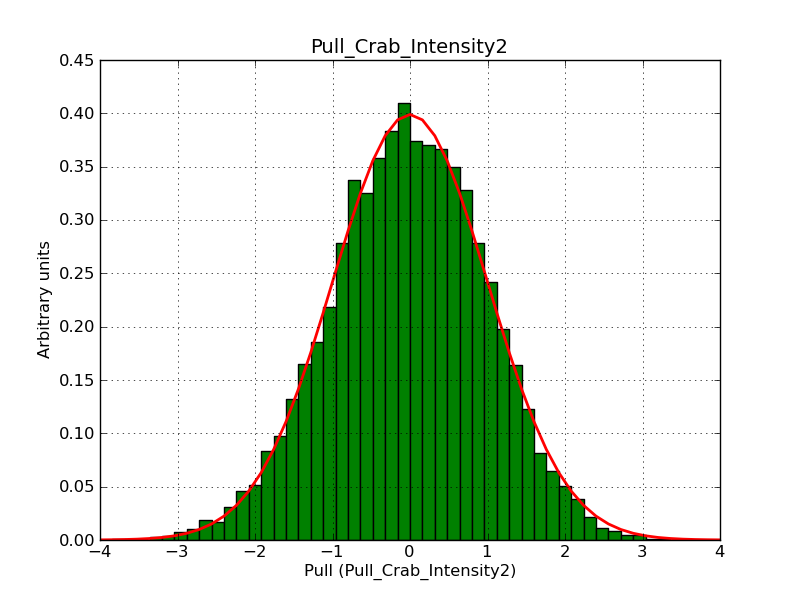
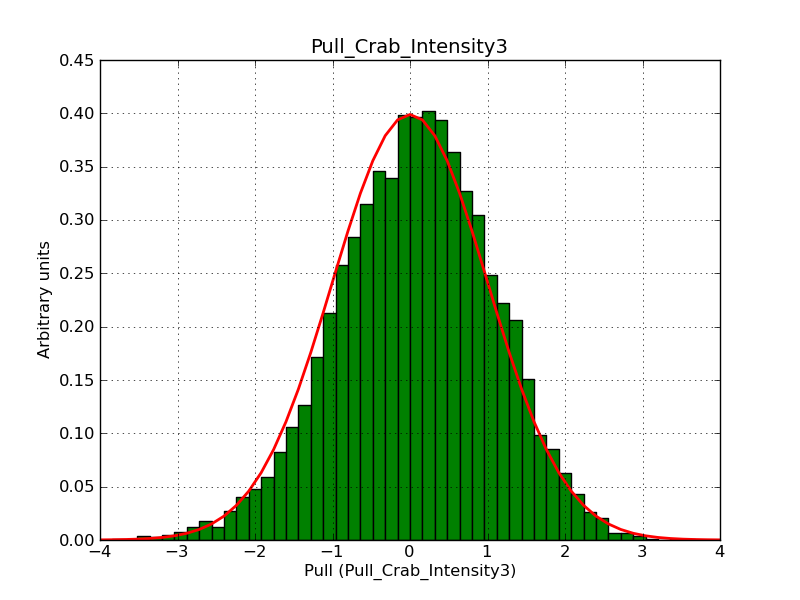
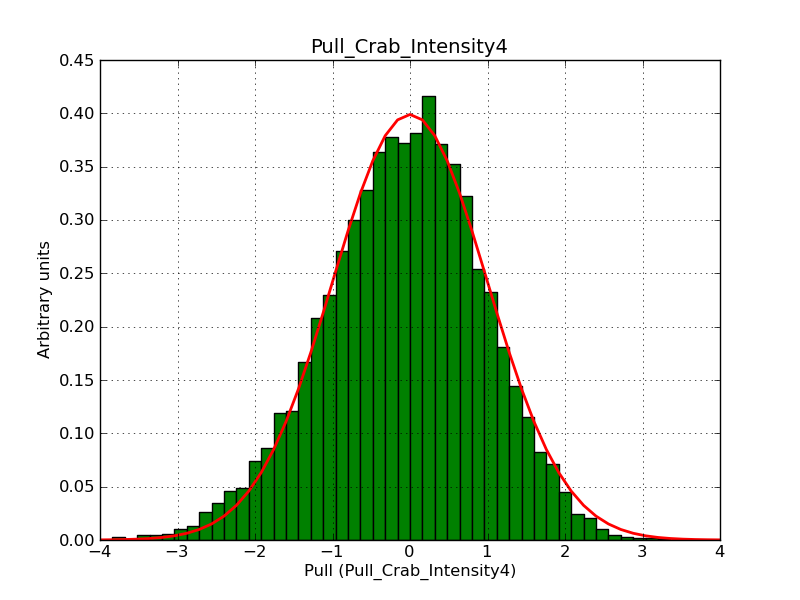
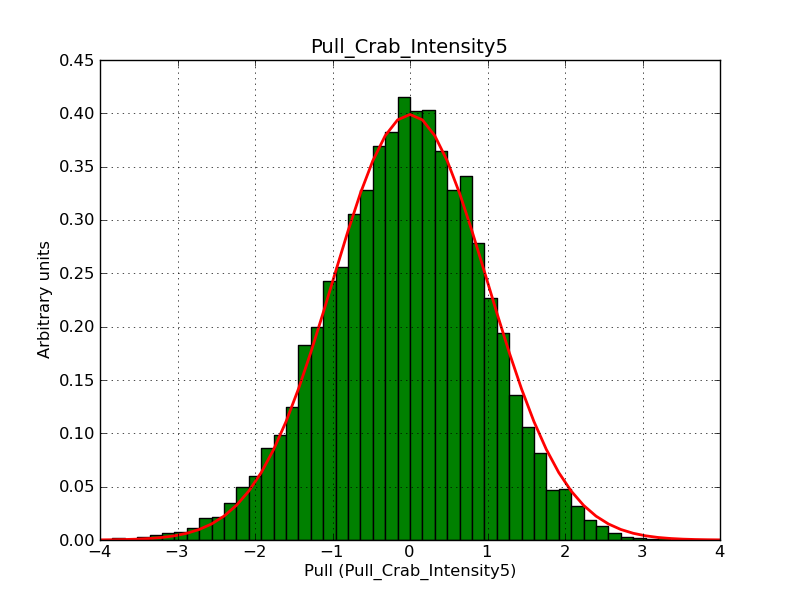
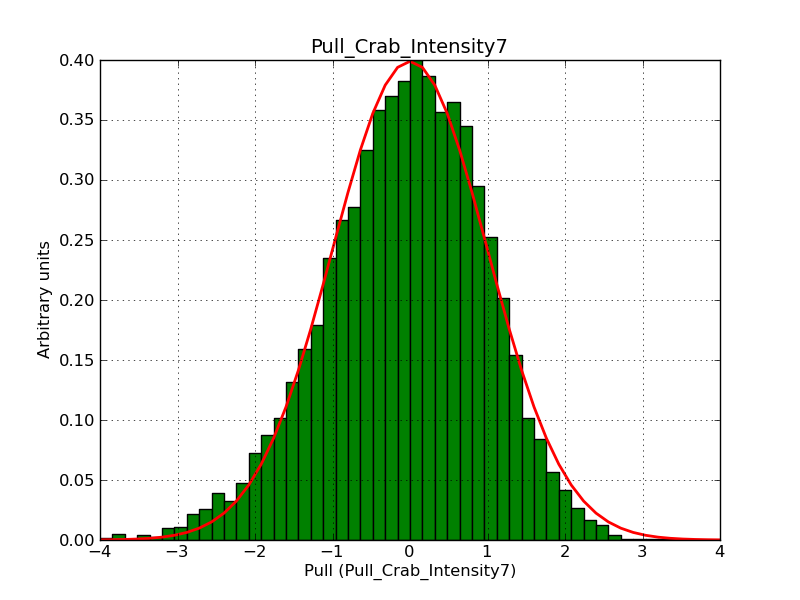
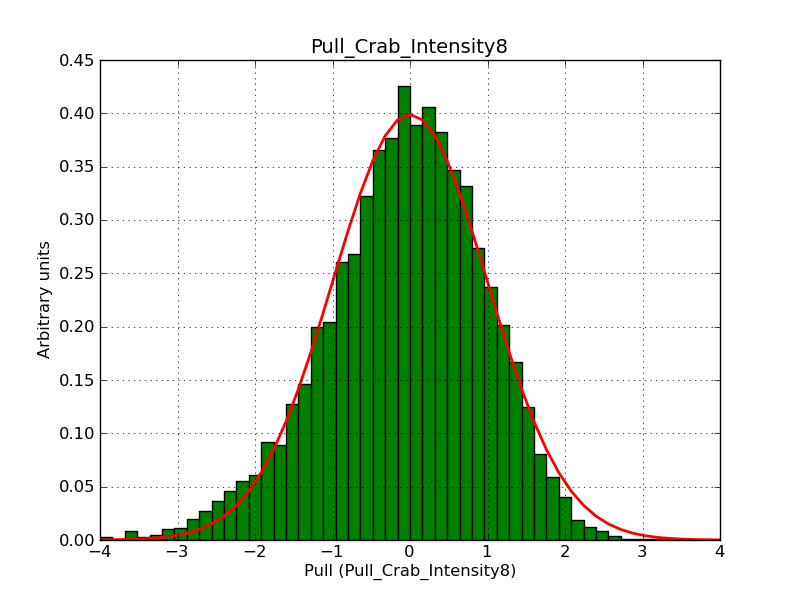
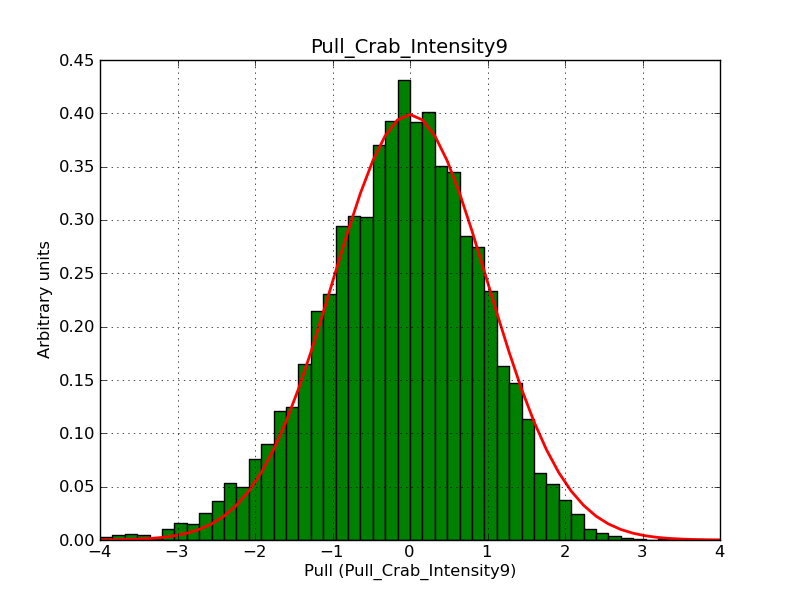
Below the pull distributions for all fitted parameters (10 spectral nodes and 2 background parameters) for 10000 Monte Carlo samples for an exposure time of 1800 seconds. The deadtime correction factor was assumed to 0.95. The file kb_E_50h_v3 was used for the instrumental response function, data have been simulated for 0.1-100 TeV for an ROI of 5 deg. The script crab_05_50_10_deadc095_t1800_n10000.sh and the model crab_05_50_10.xml was used to produce the results.
Obviously, with increasing energies the distribution becomes asymmetric. This is due to the small number of counts simulation for large energies, leading to a deviation from Gaussian statistics at high energies. Increasing the exposure time to 18000 seconds reduces the effect. The results for simulation for an exposure time of 18000 seconds are shown below.
t1800_n10000_int0.png
(44.8 KB)
Knödlseder Jürgen, 12/22/2012 09:50 PM
t1800_n10000_int1.png
(44.5 KB)
Knödlseder Jürgen, 12/22/2012 09:50 PM
t1800_n10000_int2.png
(44.9 KB)
Knödlseder Jürgen, 12/22/2012 09:50 PM
t1800_n10000_int3.png
(45 KB)
Knödlseder Jürgen, 12/22/2012 09:50 PM
t1800_n10000_int4.png
(44.7 KB)
Knödlseder Jürgen, 12/22/2012 09:50 PM
t1800_n10000_int5.png
(44.7 KB)
Knödlseder Jürgen, 12/22/2012 09:50 PM
t1800_n10000_int6.png
(39.5 KB)
Knödlseder Jürgen, 12/22/2012 09:50 PM
t1800_n10000_int7.png
(38.8 KB)
Knödlseder Jürgen, 12/22/2012 09:50 PM
t1800_n10000_int8.png
(39 KB)
Knödlseder Jürgen, 12/22/2012 09:50 PM
t1800_n10000_int9.png
(38.8 KB)
Knödlseder Jürgen, 12/22/2012 09:50 PM
t1800_n10000_bkg_norm.png
(47.7 KB)
Knödlseder Jürgen, 12/22/2012 09:51 PM
t1800_n10000_bkg_sigma.png
(46.4 KB)
Knödlseder Jürgen, 12/22/2012 09:51 PM
crab_05_50_10_deadc095_t1800_n10000.sh  (619 Bytes)
Knödlseder Jürgen, 12/22/2012 09:57 PM
(619 Bytes)
Knödlseder Jürgen, 12/22/2012 09:57 PM
crab_05_50_10.xml  (3.33 KB)
Knödlseder Jürgen, 12/22/2012 09:57 PM
(3.33 KB)
Knödlseder Jürgen, 12/22/2012 09:57 PM
t18000_n10000_bkg_norm.png
(47.8 KB)
Knödlseder Jürgen, 12/24/2012 09:51 AM
t18000_n10000_bkg_sigma.png
(46.5 KB)
Knödlseder Jürgen, 12/24/2012 09:51 AM
t18000_n10000_int0.png
(45 KB)
Knödlseder Jürgen, 12/24/2012 09:51 AM
t18000_n10000_int1.png
(44.6 KB)
Knödlseder Jürgen, 12/24/2012 09:51 AM
t18000_n10000_int2.png
(44.8 KB)
Knödlseder Jürgen, 12/24/2012 09:51 AM
t18000_n10000_int3.png
(44.7 KB)
Knödlseder Jürgen, 12/24/2012 09:51 AM
t18000_n10000_int4.png
(44.9 KB)
Knödlseder Jürgen, 12/24/2012 09:51 AM
t18000_n10000_int5.png
(44.8 KB)
Knödlseder Jürgen, 12/24/2012 09:51 AM
t18000_n10000_int6.png
(44.9 KB)
Knödlseder Jürgen, 12/24/2012 09:51 AM
t18000_n10000_int7.png
(44.8 KB)
Knödlseder Jürgen, 12/24/2012 09:51 AM
t18000_n10000_int8.png
(45.1 KB)
Knödlseder Jürgen, 12/24/2012 09:52 AM
t18000_n10000_int9.png
(45.2 KB)
Knödlseder Jürgen, 12/24/2012 09:52 AM
Also available in: PDF
HTML
TXT
Knödlseder Jürgen