Feature #1323
Write @cttsmap@
| Status: | Closed | Start date: | 09/23/2014 | |
|---|---|---|---|---|
| Priority: | Normal | Due date: | ||
| Assigned To: | % Done: | 100% | ||
| Category: | - | Estimated time: | 4.00 hours | |
| Target version: | 00-08-00 | |||
| Duration: |
Description
To statistically investigate the sky map for structures of significant gamma-ray emission, the computation of a TS map is important. Such a tool would take similar input parameters as ctbin. Thanks to the implementation of the TS calculation in ctlike (#1244), cttsmap could be rather simple.
An alternative would be to write a simple csscript instead of a new ctool. Any thoughts?
Recurrence
No recurrence.
History
#1
 Updated by Knödlseder Jürgen over 10 years ago
Updated by Knödlseder Jürgen over 10 years ago
I think I would write a specific ctool for this purpose that should be optimized for speed. Just using ctlike would not be very efficient, as you have to calculate the TS null hypothesis only once. Furthermore, we may use an optimizer without computation of the statistical parameter uncertainties, at least if one is only interested in the TS values. This would need some small modification at the GammaLib level, as the fit for the moment includes error computation. I think it would be more universal to make this a separate method.
#2
 Updated by Mayer Michael over 10 years ago
Updated by Mayer Michael over 10 years ago
I agree. Do you mean we would need to write a new GOptimizer or just to add a new method to GOptimizerLM?
The tool could also be more general, i.e. could create flux maps, maps of spectral index, etc. (anything we could get from a fit of a point source).
#3
 Updated by Knödlseder Jürgen over 10 years ago
Updated by Knödlseder Jürgen over 10 years ago
When you check the GOptimizerLM::optimize method you’ll see at the end a call to the errors method to compute parameter errors. We could in principle drop this call and call it explicitly in ctlike. ctlike calls actually GObservations::optimize, which has the following code:
void GObservations::optimize(GOptimizer& opt)
{
// Extract optimizer parameter container from model container
GOptimizerPars pars = m_models.pars();
// Optimize model parameters
opt.optimize(m_fct, pars);
// Return
return;
}
We could add a method
GObservations::errors:void GObservations::errors(GOptimizer& opt)
{
// Extract optimizer parameter container from model container
GOptimizerPars pars = m_models.pars();
// Compute model parameter errors
opt.errors(m_fct, pars);
// Return
return;
}
that simply calls the optimizer’s
errors method. So far, errors is not pure virtual in GOptimizer, hence this method needs to be added to the interface.
In ctlike, we then would need to change ctlike::optimize_lm as follows:
// Perform LM optimization
m_obs.optimize(*opt);
// Optionally refit
if (m_refit) {
m_obs.optimize(*opt);
}
// Compute errors
m_obs.errors(*opt);
The advantage would also be that in ctlike::reoptimize_lm no error computations are done, hence TS computation should be quicker.
#4
 Updated by Mayer Michael over 10 years ago
Updated by Mayer Michael over 10 years ago
- Assigned To set to Mayer Michael
- Target version set to 00-08-00
- % Done changed from 0 to 30
- Estimated time set to 4.00
Great, I thought of another, similar solution by adding a flag to GOptimizer::optimize(..., bool errors). This flag could be set to true on default in the base class and only gets dropped for the TS calculations.
But I guess your suggestion is more general. What is your preference?
I am currently writing the cttsmap-tool.
#5
 Updated by Knödlseder Jürgen over 10 years ago
Updated by Knödlseder Jürgen over 10 years ago
I thought also about a flag, but this makes things a little more opaque. I think it´s cleaner to separate optimization from error computation. This would even give the possibility to have different methods to compute the error (we know e.g. that the actual method has problems with correlated errors). Maybe we could even split the error computation part completely from the optimizer part. Strictly speaken, both have nothing to do with each other.
#6
 Updated by Mayer Michael over 10 years ago
Updated by Mayer Michael over 10 years ago
- Status changed from New to Feedback
- % Done changed from 30 to 90
I agree and made issue #1328 for the changes in gammalib.
The first version of cttsmap is now available on branch 1323-write-tsmap.
- The user should be able to provide minimum, maximum and start values for the parameters Integral and Index for the putative point source because they might change for Fermi and IACTs. Another option would be to store these in an xml-file, which would give the user access also to the spectral model and I don’t know if this is desired.
- The parameter free_index allows the user to control wether the spectral index of the test source should be fixed.
- I’ve added the parameters binmin and binmax to just compute a part of the TS map. In case the computing time for one bin is quite long, the user can split the calculations to different jobs. Of course, we would need to write
cttsmerge, which, however, might be not that difficult. - In case of job splitting, one might pass the likelihood value of the null hypothesis, such that this doesn’t have to be recomputed for every single job.
#7
 Updated by Knödlseder Jürgen over 10 years ago
Updated by Knödlseder Jürgen over 10 years ago
I looked into the cttsmap parameter file. I would propose to add simply a “test sources” to the XML file, which then would allow to specify initial parameters and boundaries in the XML file. The only parameter needed would then be the source name, and cttsmap should check whether the source exists, and whether it has a RA and DEC parameter that will be needed for building the map. This would also to have the advantage that we could make TS maps for extended sources, e.g. Gaussians where for each position the width is fitted.
#8
 Updated by Mayer Michael over 10 years ago
Updated by Mayer Michael over 10 years ago
- Status changed from Feedback to Pull request
- % Done changed from 90 to 100
Good idea. I changed the par file and the code accordingly. Now, the user has to specify a source name in the xml model, which is moved around the coordinate grid. cttsmap checks for RA and DEC parameters of the source and uses the given spectral model.
#9
 Updated by Knödlseder Jürgen over 10 years ago
Updated by Knödlseder Jürgen over 10 years ago
- Status changed from Pull request to Closed
Thanks Michael. This is really a nice tool.
I did some minor modifications during integration:- change evfile parameter to infile (as the file will also work on counts cubes, hence evfile is misleading)
- added irf and caldb parameters (to specify the calibration database and IRF when running from then command line)
- use pointer to GModel for test source (this avoids dynamic casting to GModelSky)
- clean exception handling (use now invalid_value() throughout)
- minor code adjustments (for example in the logging)
I also unit tests.
Is now merged into devel.
I now also understand your binmin and binmax parameters. This is for splitting manually the computation in several junks. I guess you have then some scripts for merging?
Somehow the word bin was misleading to me, as I was thinking about the data space. Maybe grid would be more appropriate? The tool would be ideal also for parallelization using OpenMP. This is a little tricky as one has to avoid bottlenecks from protecting some memory. We can keep this as a possible future option. Would parallelization avoid the need having the binmin and binmax parameters?
#10
 Updated by Knödlseder Jürgen over 10 years ago
Updated by Knödlseder Jürgen over 10 years ago
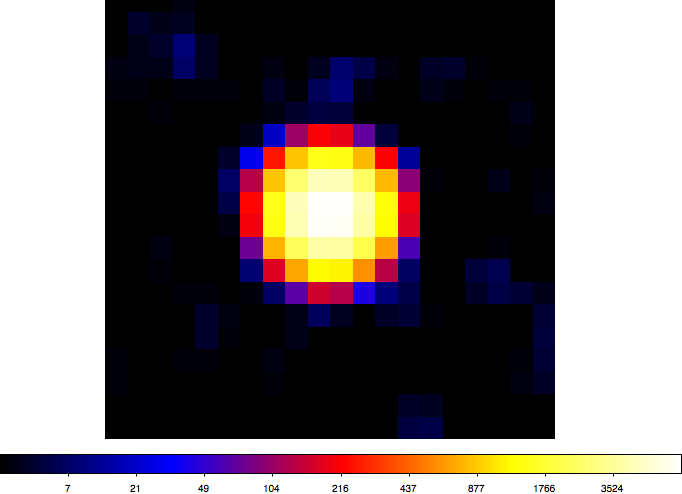
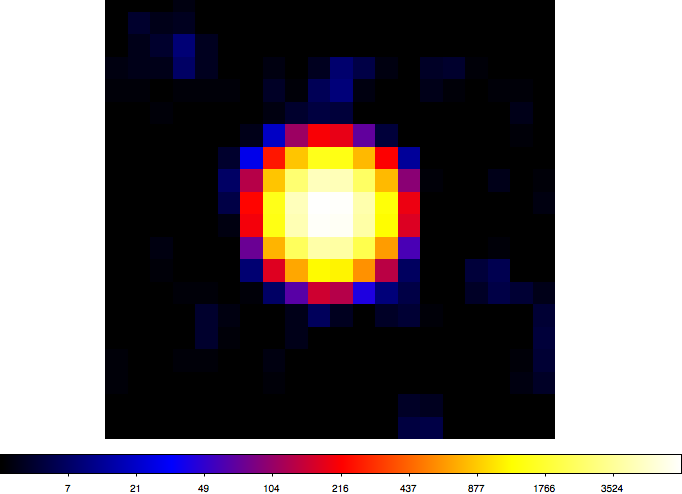
- File crab-ts.png added
Here a TS map of the Crab test data for a 1°x1° field with 0.05° grid spacing (20x20 grid points). Computation took less than 2 minutes on my Mac OS X 10.6 2.66 GHz Intel Core i7 (image is in logarithmic scaling).
#11
 Updated by Mayer Michael over 10 years ago
Updated by Mayer Michael over 10 years ago
looks good! Thanks for making the adjustments. I have to look deeper into the logger to understand what is going on and next time take that into account when writing the code.
I guess you have then some scripts for merging?
I was thinking about writing cttsmerge, which takes a list of files to merge as an argument. Does that sound like a reasonable approach?
Somehow the word bin was misleading to me, as I was thinking about the data space. Maybe grid would be more appropriate?
I agree that gridmin and gridmax might be more meaningful. Still, it doesn’t seem like an ideal solution to me. Somehting like compute_first and compute_last could be more descriptive.
The tool would be ideal also for parallelization using OpenMP. This is a little tricky as one has to avoid bottlenecks from protecting some memory. We can keep this as a possible future option. Would parallelization avoid the need having the binmin and binmax parameters?
I am not sure. If OpenMP is running on the system, the model optimisation is anyhow distributed on several cores. I don’t know if we would gain from computing each bin on one core instead of using multi cores for the individual optimisation steps. Especially, when the dataset becomes large and the models are complex, i.e. extended or diffuse, the user might even want to calculate one bin per job. Having a fine-binned map with 100x100 bins, could then be splitted into 1e4 jobs, each using a reasonable amount of computing time.
#12
 Updated by Knödlseder Jürgen over 10 years ago
Updated by Knödlseder Jürgen over 10 years ago
Mayer Michael wrote:
looks good! Thanks for making the adjustments. I have to look deeper into the logger to understand what is going on and next time take that into account when writing the code.
I guess you have then some scripts for merging?
I was thinking about writingcttsmerge, which takes a list of files to merge as an argument. Does that sound like a reasonable approach?
Looks a bit heavy for what is needed here. I guess I would do this really at the script level, as this allows more flexibility. One may also do the handling of the jobs on script level, that’s by the way how I actually do a TS map for Fermi (run one gtlike job per position and use a script to launch the jobs and collect the results).
Still, would be good if ctools could be more clever.
Somehow the word bin was misleading to me, as I was thinking about the data space. Maybe grid would be more appropriate?
I agree that gridmin and gridmax might be more meaningful. Still, it doesn’t seem like an ideal solution to me. Somehting like compute_first and compute_last could be more descriptive.The tool would be ideal also for parallelization using OpenMP. This is a little tricky as one has to avoid bottlenecks from protecting some memory. We can keep this as a possible future option. Would parallelization avoid the need having the binmin and binmax parameters?
I am not sure. If OpenMP is running on the system, the model optimisation is anyhow distributed on several cores. I don’t know if we would gain from computing each bin on one core instead of using multi cores for the individual optimisation steps. Especially, when the dataset becomes large and the models are complex, i.e. extended or diffuse, the user might even want to calculate one bin per job. Having a fine-binned map with 100x100 bins, could then be splitted into 1e4 jobs, each using a reasonable amount of computing time.
OpenMP is for the moment only used if there a multiple observations in an XML file. For cube-style analysis there won’t be any benefit so far. So I guess it would make sense to implement OpenMP support here. We also have to keep in mind that the number of cores per CPU will likely evolve in the future, hence having a few dozen cores in the future on your laptop is probably not impossible.
But I agree that the user may want some more fine tuning capabilities.
#13
 Updated by Mayer Michael over 10 years ago
Updated by Mayer Michael over 10 years ago
Knödlseder Jürgen wrote:
Looks a bit heavy for what is needed here. I guess I would do this really at the script level, as this allows more flexibility. One may also do the handling of the jobs on script level, that’s by the way how I actually do a TS map for Fermi (run one gtlike job per position and use a script to launch the jobs and collect the results).
Before cttsmap, I also wrote the results of each bin in an ascii-file. All files were merged into a sky map at the end. For cttsmap, I guess it is better to stick to FITS files in order not to introduce a new ascii file format. I agree that a ctool for merging might be a bit bulky for this simple job. Do you mean to write a csscript then, or just having a simple script somewhere else?
One might also think about an option for cttsmap, e.g calling it with
cttsmap merge=yes
will query for the file list and merges the files immediately.
OpenMP is for the moment only used if there a multiple observations in an XML file. For cube-style analysis there won’t be any benefit so far. So I guess it would make sense to implement OpenMP support here. We also have to keep in mind that the number of cores per CPU will likely evolve in the future, hence having a few dozen cores in the future on your laptop is probably not impossible.
But I agree that the user may want some more fine tuning capabilities.
I guess with cube-style analysis you mean binned analysis, right? For this side, it makes sense to provide OpenMP support. However, for unbinned analysis (which I currently apply), it might get quite complex. OpenMP would have to steer jobs, which in turn get parallelised. Do you think this is possible?
#14
 Updated by Knödlseder Jürgen over 10 years ago
Updated by Knödlseder Jürgen over 10 years ago
I was thinking about a simple script, at the obsutils level. But we may just see how things evolve and how the cttsmap tool will be used in the future to get a better feeling for what is needed.
Concerning OpenMP, I expect that the system deals automatically with nested parallelization, but this remains to be investigated. It’s not an urgent issue, but something we may want to have on the long run.
#15
 Updated by Mayer Michael over 10 years ago
Updated by Mayer Michael over 10 years ago
- File tsmerge.py
 added
added
I attach a script to merge the output files of cttsmap (I currently cannot push it into the branch for some reasons, as I am on travel). The script could be located in "$CTOOLS/scripts/”. The usage of the script is (hopefully) simple enough:
python tsmerge.py files=<list of files> outfile=<name of output file>
<list of files> could be a wildcard string, or an ascii-file containing the file names.
Concerning OpenMP, I expect that the system deals automatically with nested parallelization, but this remains to be investigated. It’s not an urgent issue, but something we may want to have on the long run.
If nested parallelisations are dealed with automatically, it would be nice to have the option available at some point. We could make an issue for it, that we don’t forget.
#16
 Updated by Knödlseder Jürgen over 10 years ago
Updated by Knödlseder Jürgen over 10 years ago
Thanks Michael, I merged the script into the devel branch. I have done no testing, though.
#17
 Updated by Mayer Michael over 10 years ago
Updated by Mayer Michael over 10 years ago
I tested the script for a tsmap, which was splitted into 4 jobs and merged it afterwards. The results look satisfactory. Let’s hope it stays like this