Feature #1991
Source Probability in event file - ctprob
| Status: | Closed | Start date: | 04/11/2017 | |
|---|---|---|---|---|
| Priority: | Normal | Due date: | ||
| Assigned To: | % Done: | 100% | ||
| Category: | - | Estimated time: | 6.00 hours | |
| Target version: | 1.3.0 | |||
| Duration: |
Description
Create new ctool: ctprob
The tool calculates the probability for each photon in the input event list to come from each of the sources in a given model
Recurrence
No recurrence.
History
#1
 Updated by Di Venere Leonardo almost 8 years ago
Updated by Di Venere Leonardo almost 8 years ago
- Tracker changed from Bug to Feature
- Start date deleted (
04/06/2017)
#2
 Updated by Di Venere Leonardo almost 8 years ago
Updated by Di Venere Leonardo almost 8 years ago
- File probabilities1.png added
- File probabilities2.png added
- Status changed from New to Pull request
- % Done changed from 0 to 100
The tool ctprob has been developed and tested.
The tool takes an event list file (or an XML observation file) and a source model as input.
For each event, it evalutes the probability for that event to come from each of the source in the model.
A column for each source in the model is added to the event list in order to store these values.
In order to test the tool, I generate an event list, simulating a crab spectrum and the background using ctobssim.
Then I added an additional source in the model used in ctprob and tested the tool in two cases.
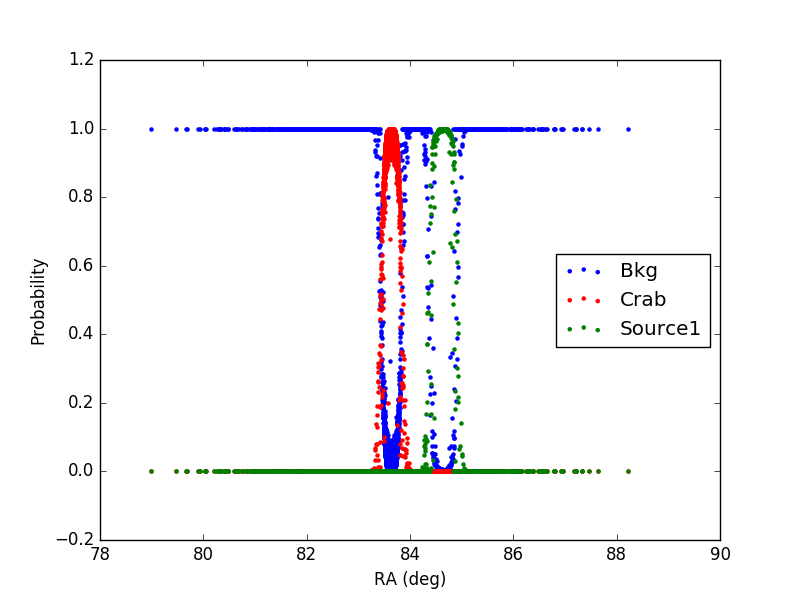
1) In the first case, the source (Source1) has a flux 10 times larger than the Crab and an offset of 1 degree from the Crab (coords_source1 = (RA_crab+1, DEC_crab) )
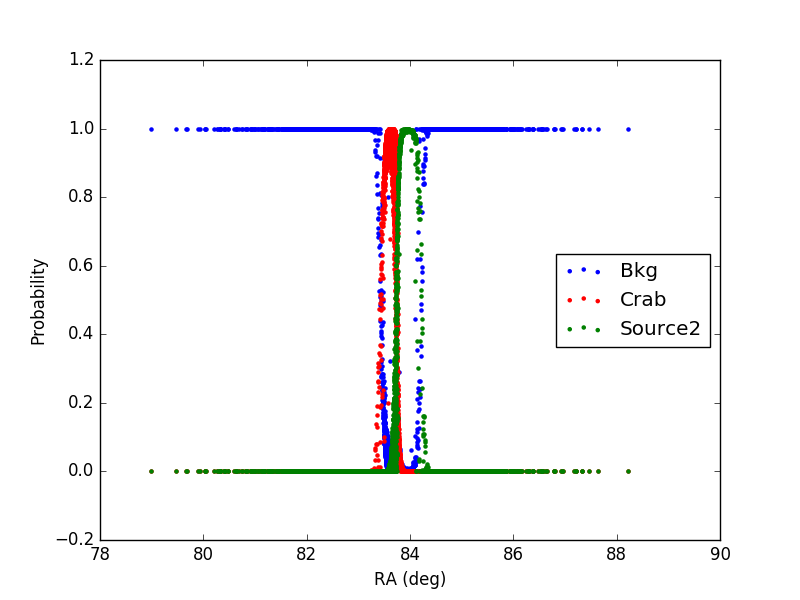
2) In the second case, the source (Source2) has a flux 10 times larger than the Crab (same as in case 1) and an offset of 0.3 degree from the Crab (coords_source2 = (RA_crab+0.3, DEC_crab) )
In the plot, only events with declination in the interval (DEC_crab-0.1, DEC_crab+0.1) are included.
It can be noticed that for each event the sum of the probabilities is 1 as expected.
Furthermore, the additional source (1 or 2) has a larger distribution with respect to the Crab, being the flux larger.
Finally, in case 2, in the region in-between the Crab and Source2 the probability is correctly shared between the two sources, according to the distance from the source and its flux.
The tool seems to be working properly.
I also added a few python tests, one of which takes an observation XML file as an input.
-----
NOTE:
A pull of the associated gammaLib branch is also necessary. In particular, I added a new method to the GCTAEventList class that appends a new column to “m_columns”. This is necessary to correctly add the new columns containing the source probabilities to the event list and then save them to the output file.
-----
The relevant branches are:
- ctools: 1991-ctprob-eval-event-probs
- gammalib: 1991-ctprob-modified-
Figure1: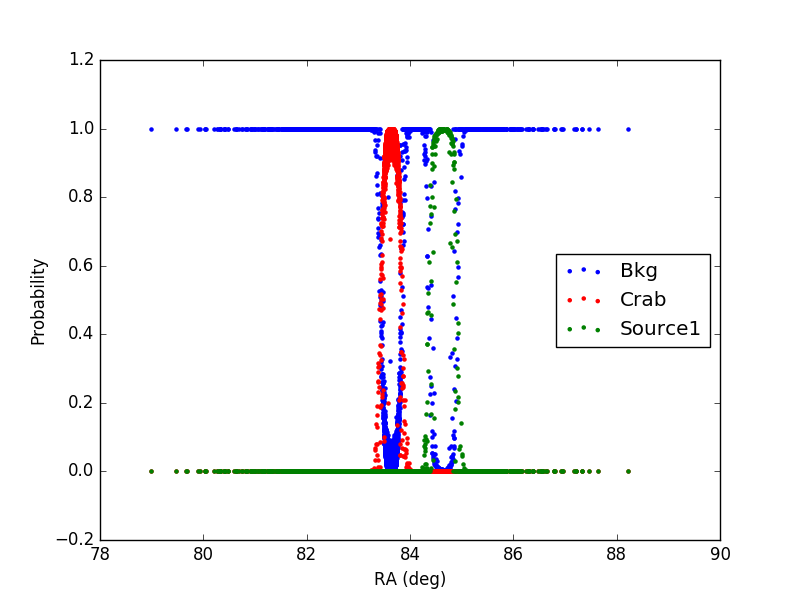
Figure2: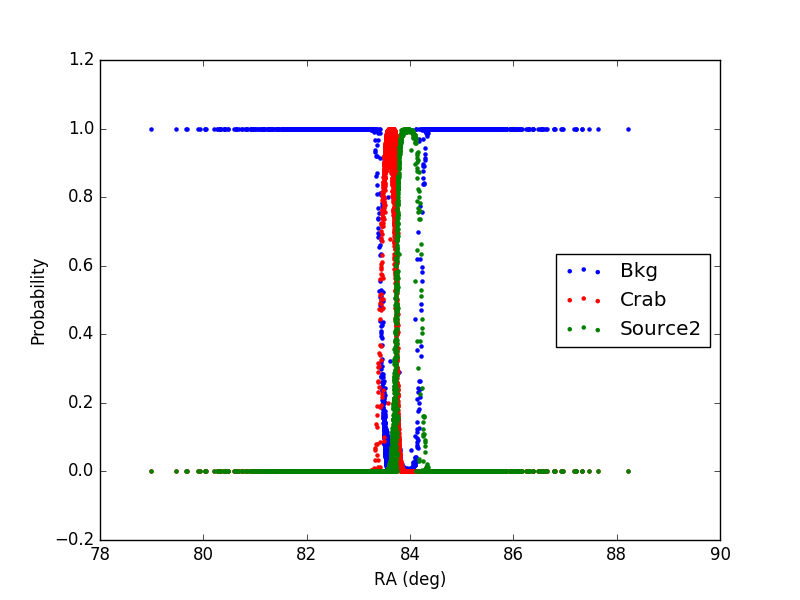
#3
 Updated by Knödlseder Jürgen almost 8 years ago
Updated by Knödlseder Jürgen almost 8 years ago
- Target version set to 1.3.0
- Start date set to 04/11/2017
I integrated the GammaLib part. Note that the GCTAEventList::append_column() method as written was vulnerable to a memory leak since it appended the reference to the column provided on input to the GCTAEventList object. Since the memory has been created outside the method the GCTAEventList class has no control over when the memory is de-allocated.
I changed the code so that a clone of the column is appended. Now, GCTAEventList has full control over the memory allocation and de-allocation, and the client can do what he wants with the FITS column.
I also added an exception in case that the column length is incompatible with the number of events.
#4
 Updated by Knödlseder Jürgen almost 8 years ago
Updated by Knödlseder Jürgen almost 8 years ago
- Status changed from Pull request to Closed
I now merged the ctprob tool into ctools.
I refactored a bit the code. For example the writing of a temporary FITS file is not needed. I also put some code that is redundant with ctselect in the src/support/ctool.cpp file.
All code is in devel now.