Bug #2683
Errors with On/Off analysis of HESS data in stacked mode
| Status: | Closed | Start date: | 09/20/2018 | |
|---|---|---|---|---|
| Priority: | Normal | Due date: | ||
| Assigned To: | % Done: | 100% | ||
| Category: | - | |||
| Target version: | 1.6.0 | |||
| Duration: |
Description
I found some issues in an analysis of the public HESS data using the On/Off analysis in stacked mode.
I used the selected Crab observations prepared following this tutorial
http://cta.irap.omp.eu/ctools-devel/users/tutorials/hess_dr1/select_events.html
Then I created stacked On/Off observations doing
onoff_obs = 'obs_crab_onoff_stacked.xml' phagen = cscripts.csphagen() phagen['inobs'] = 'obs_crab_selected.xml' phagen['inmodel'] = 'crab_models.xml' phagen['srcname'] = 'Crab' phagen['outobs'] = onoff_obs phagen['ebinalg'] = 'LOG' phagen['emin'] = 0.66 phagen['emax'] = 50. phagen['enumbins'] = 12 phagen['coordsys'] = 'CEL' phagen['ra'] = 83.63 phagen['dec'] = 22.01 phagen['rad'] = 0.2 phagen['bkgmethod'] = 'REFLECTED' phagen['use_irf_bkg'] = False phagen['maxoffset'] = 2.0 phagen['stack'] = True phagen.execute()
If I try to run ctlike on these On/Off observations I get
(ctools) macp0077:tmp ltibaldo$ ctlike statistic=wstat debug=yes chatter=4 Input event list, counts cube or observation definition XML file [obs_crab_onoff.xml] obs_crab_onoff_stacked.xml Input model definition XML file [crab_models.xml] Output model definition XML file [crab_plaw_stacked.xml] 2018-09-20T09:35:12: +============+ 2018-09-20T09:35:12: | Parameters | 2018-09-20T09:35:12: +============+ 2018-09-20T09:35:12: inobs .....................: obs_crab_onoff_stacked.xml 2018-09-20T09:35:12: inmodel ...................: crab_models.xml 2018-09-20T09:35:12: expcube ...................: [not queried] 2018-09-20T09:35:12: psfcube ...................: [not queried] 2018-09-20T09:35:12: edispcube .................: [not queried] 2018-09-20T09:35:12: bkgcube ...................: [not queried] 2018-09-20T09:35:12: caldb .....................: [not queried] 2018-09-20T09:35:12: irf .......................: [not queried] 2018-09-20T09:35:12: edisp .....................: no 2018-09-20T09:35:12: outmodel ..................: crab_plaw_stacked.xml 2018-09-20T09:35:12: outcovmat .................: NONE 2018-09-20T09:35:12: statistic .................: wstat 2018-09-20T09:35:12: refit .....................: no 2018-09-20T09:35:12: like_accuracy .............: 0.005 2018-09-20T09:35:12: fix_spat_for_ts ...........: no 2018-09-20T09:35:12: nthreads ..................: 0 2018-09-20T09:35:12: chatter ...................: 4 2018-09-20T09:35:12: clobber ...................: yes 2018-09-20T09:35:12: debug .....................: yes 2018-09-20T09:35:12: mode ......................: ql 2018-09-20T09:35:12: logfile ...................: ctlike.log 2018-09-20T09:35:12: 2018-09-20T09:35:12: +===================+ 2018-09-20T09:35:12: | Input observation | 2018-09-20T09:35:12: +===================+ 2018-09-20T09:35:12: === GObservations === 2018-09-20T09:35:12: Number of observations ....: 1 2018-09-20T09:35:12: Number of models ..........: 1 2018-09-20T09:35:12: Number of observed events .: 534 2018-09-20T09:35:12: Number of predicted events : 0 2018-09-20T09:35:12: === GCTAOnOffObservation === 2018-09-20T09:35:12: Name ......................: 2018-09-20T09:35:12: Identifier ................: 2018-09-20T09:35:12: Instrument ................: CTAOnOff 2018-09-20T09:35:12: Statistic .................: WSTAT 2018-09-20T09:35:12: Ontime ....................: 1581.7368164 s 2018-09-20T09:35:12: Livetime ..................: 1581.7368164 s 2018-09-20T09:35:12: Deadtime correction .......: 1 2018-09-20T09:35:12: === GPha === 2018-09-20T09:35:12: Number of bins ............: 12 2018-09-20T09:35:12: Energy range ..............: 660 GeV - 50 TeV 2018-09-20T09:35:12: Observation energy range ..: 870.9635625 GeV - 100 TeV 2018-09-20T09:35:12: Total number of counts ....: 534 2018-09-20T09:35:12: Underflow counts ..........: 0 2018-09-20T09:35:12: Overflow counts ...........: 0 2018-09-20T09:35:12: Outflow counts ............: 0 2018-09-20T09:35:12: === GPha === 2018-09-20T09:35:12: Number of bins ............: 12 2018-09-20T09:35:12: Energy range ..............: 660 GeV - 50 TeV 2018-09-20T09:35:12: Observation energy range ..: 870.9635625 GeV - 100 TeV 2018-09-20T09:35:12: Total number of counts ....: 706 2018-09-20T09:35:12: Underflow counts ..........: 0 2018-09-20T09:35:12: Overflow counts ...........: 0 2018-09-20T09:35:12: Outflow counts ............: 0 2018-09-20T09:35:12: === GArf === 2018-09-20T09:35:12: Number of bins ............: 68 2018-09-20T09:35:12: Energy range ..............: 330 GeV - 60.000002048 TeV 2018-09-20T09:35:12: === GRmf === 2018-09-20T09:35:12: Number of true energy bins : 68 2018-09-20T09:35:12: Number of measured bins ...: 12 2018-09-20T09:35:12: True energy range .........: 330 GeV - 60.000002048 TeV 2018-09-20T09:35:12: Measured energy range .....: 660 GeV - 50 TeV 2018-09-20T09:35:12: === GModels === 2018-09-20T09:35:12: Number of models ..........: 1 2018-09-20T09:35:12: Number of parameters ......: 6 2018-09-20T09:35:12: === GModelSky === 2018-09-20T09:35:12: Name ......................: Crab 2018-09-20T09:35:12: Instruments ...............: all 2018-09-20T09:35:12: Instrument scale factors ..: unity 2018-09-20T09:35:12: Observation identifiers ...: all 2018-09-20T09:35:12: Model type ................: PointSource 2018-09-20T09:35:12: Model components ..........: "PointSource" * "PowerLaw" * "Constant" 2018-09-20T09:35:12: Number of parameters ......: 6 2018-09-20T09:35:12: Number of spatial par's ...: 2 2018-09-20T09:35:12: RA .......................: 83.63 deg (fixed,scale=1) 2018-09-20T09:35:12: DEC ......................: 22.01 deg (fixed,scale=1) 2018-09-20T09:35:12: Number of spectral par's ..: 3 2018-09-20T09:35:12: Prefactor ................: 5.7e-17 +/- 0 [0,infty[ ph/cm2/s/MeV (free,scale=1e-17,gradient) 2018-09-20T09:35:12: Index ....................: -2.5 +/- 0 [4,-4] (free,scale=-1,gradient) 2018-09-20T09:35:12: PivotEnergy ..............: 1000000 MeV (fixed,scale=1000000,gradient) 2018-09-20T09:35:12: Number of temporal par's ..: 1 2018-09-20T09:35:12: Normalization ............: 1 (relative value) (fixed,scale=1,gradient) 2018-09-20T09:35:12: 2018-09-20T09:35:12: +=================================+ 2018-09-20T09:35:12: | Maximum likelihood optimisation | 2018-09-20T09:35:12: +=================================+ 2018-09-20T09:35:12: >Iteration 0: -logL=nan, Lambda=1.0e-03 *** ERROR: GModelSpectralPlaw::eval(srcEng=682.650765402332 GeV, srcTime=0 s (TT)): NaN/Inf encountered (value=nan, m_norm=nan, m_index=nan, m_pivot=1e+06, m_last_power=nan) *** ERROR: GModelSpectralPlaw::eval(srcEng=736.93388421952 GeV, srcTime=0 s (TT)): NaN/Inf encountered (value=nan, m_norm=nan, m_index=nan, m_pivot=1e+06, m_last_power=nan) *** ERROR: GModelSpectralPlaw::eval(srcEng=795.533521124894 GeV, srcTime=0 s (TT)): NaN/Inf encountered (value=nan, m_norm=nan, m_index=nan, m_pivot=1e+06, m_last_power=nan) ...
The joint On/Off analysis, on the other hand, works as expected. If I calculate residuals using csresspec this works as expected when not stacking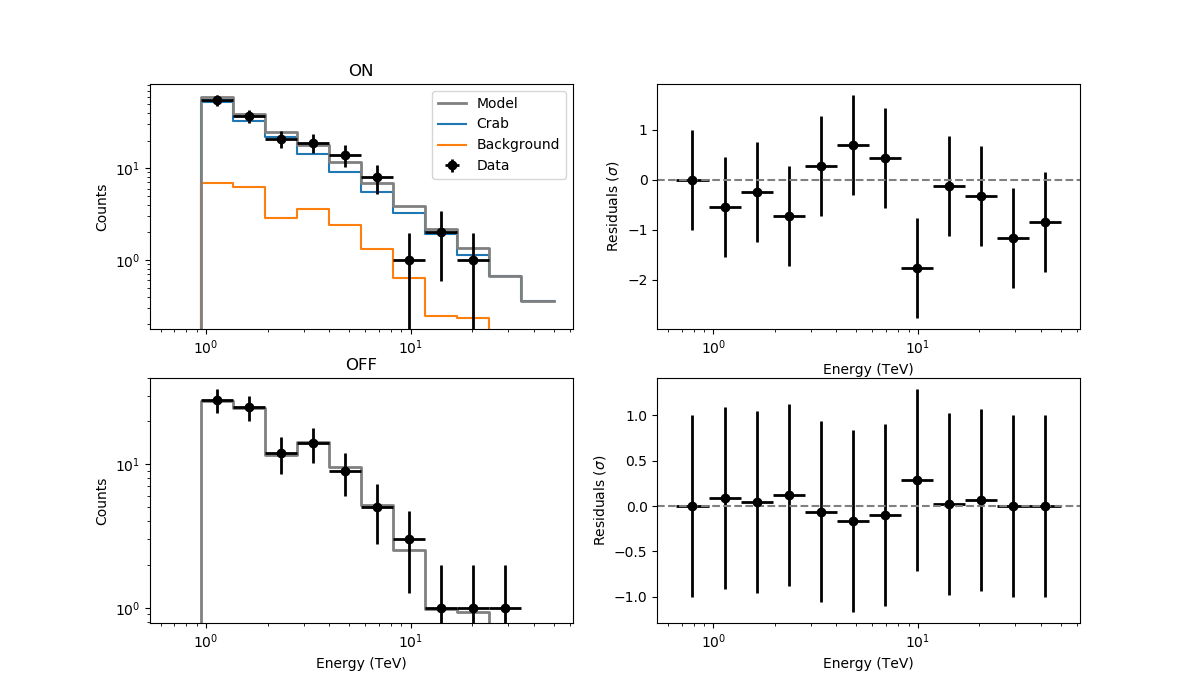
but stacked residuals don’t, the background is always set to 0.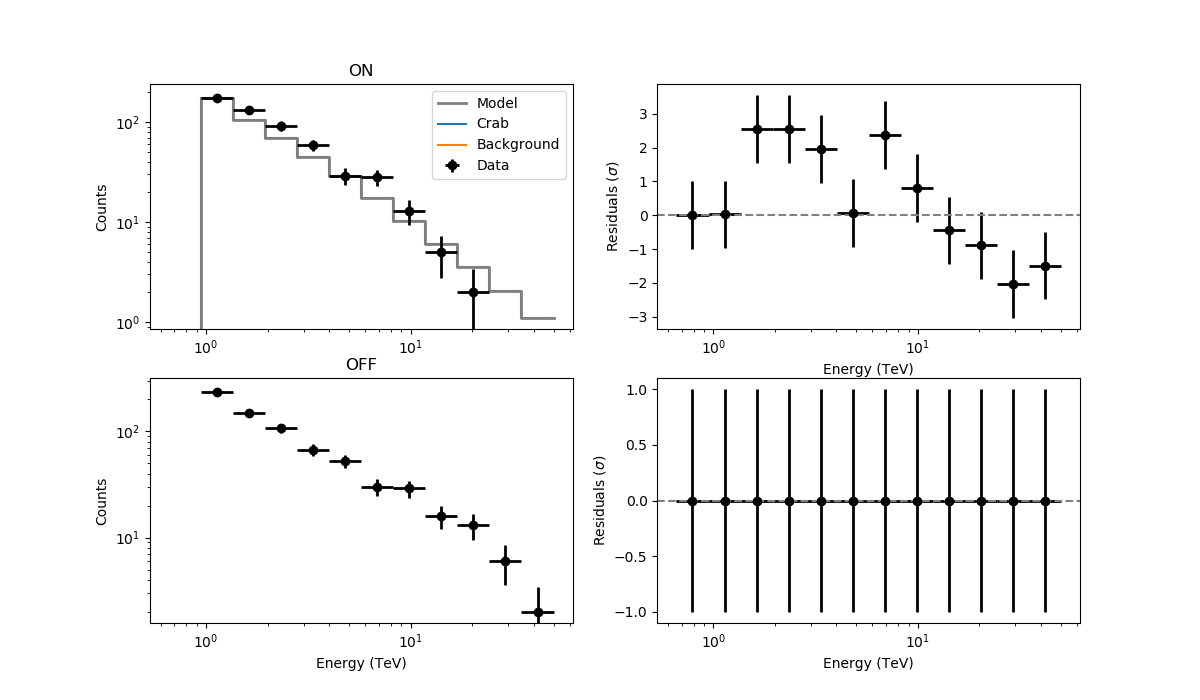
I guess this may be related to the absence of a background IRF template, or to stacking with varying energy thresholds.
Recurrence
No recurrence.
Related issues
History
#1
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
- Status changed from New to Pull request
- Assigned To set to Knödlseder Jürgen
- Target version set to 1.6.0
- % Done changed from 0 to 100
The issue came from GCTAOnOffObservation(const GObservations& obs) which did not correctly handle the case where the IRF background template is absent. I modified the code so that the handling is now done correctly.
Here is the ctlike result after having done the change:
2018-10-04T08:33:36: +=================================+ 2018-10-04T08:33:36: | Maximum likelihood optimisation | 2018-10-04T08:33:36: +=================================+ 2018-10-04T08:33:36: >Iteration 0: -logL=209.552, Lambda=1.0e-03 2018-10-04T08:33:36: >Iteration 1: -logL=97.156, Lambda=1.0e-03, delta=112.396, step=1.0e+00, max(|grad|)=266.954813 [Index:3] 2018-10-04T08:33:36: >Iteration 2: -logL=14.417, Lambda=1.0e-04, delta=82.740, step=1.0e+00, max(|grad|)=69.552529 [Index:3] 2018-10-04T08:33:36: >Iteration 3: -logL=11.287, Lambda=1.0e-05, delta=3.130, step=1.0e+00, max(|grad|)=41.225603 [Index:3] 2018-10-04T08:33:36: >Iteration 4: -logL=10.609, Lambda=1.0e-06, delta=0.678, step=1.0e+00, max(|grad|)=-0.660643 [Index:3] 2018-10-04T08:33:36: >Iteration 5: -logL=10.604, Lambda=1.0e-07, delta=0.005, step=1.0e+00, max(|grad|)=0.563913 [Index:3] 2018-10-04T08:33:36: 2018-10-04T08:33:36: +==================+ 2018-10-04T08:33:36: | Curvature matrix | 2018-10-04T08:33:36: +==================+ 2018-10-04T08:33:36: === GMatrixSparse === 2018-10-04T08:33:36: Number of rows ............: 6 2018-10-04T08:33:36: Number of columns .........: 6 2018-10-04T08:33:36: Number of nonzero elements : 4 2018-10-04T08:33:36: Number of allocated cells .: 516 2018-10-04T08:33:36: Memory block size .........: 512 2018-10-04T08:33:36: Sparse matrix fill ........: 0.111111111111111 2018-10-04T08:33:36: Pending element ...........: 0 2018-10-04T08:33:36: Fill stack size ...........: 0 (none) 2018-10-04T08:33:36: 0, 0, 0, 0, 0, 0 2018-10-04T08:33:36: 0, 0, 0, 0, 0, 0 2018-10-04T08:33:36: 0, 0, 0.152622527932884, -13.5947132096962, 0, 0 2018-10-04T08:33:36: 0, 0, -13.5947132096962, 1313.28746416818, 0, 0 2018-10-04T08:33:36: 0, 0, 0, 0, 0, 0 2018-10-04T08:33:36: 0, 0, 0, 0, 0, 0 2018-10-04T08:33:36: 2018-10-04T08:33:36: +=========================================+ 2018-10-04T08:33:36: | Maximum likelihood optimisation results | 2018-10-04T08:33:36: +=========================================+ 2018-10-04T08:33:36: === GOptimizerLM === 2018-10-04T08:33:36: Optimized function value ..: 10.604 2018-10-04T08:33:36: Absolute precision ........: 0.005 2018-10-04T08:33:36: Acceptable value decrease .: 2 2018-10-04T08:33:36: Optimization status .......: converged 2018-10-04T08:33:36: Number of parameters ......: 6 2018-10-04T08:33:36: Number of free parameters .: 2 2018-10-04T08:33:36: Number of iterations ......: 5 2018-10-04T08:33:36: Lambda ....................: 1e-08 2018-10-04T08:33:36: Maximum log likelihood ....: -10.604 2018-10-04T08:33:36: Observed events (Nobs) ...: 534.000 2018-10-04T08:33:36: Predicted events (Npred) ..: 534.505 (Nobs - Npred = -0.504815279792638) 2018-10-04T08:33:36: === GModels === 2018-10-04T08:33:36: Number of models ..........: 1 2018-10-04T08:33:36: Number of parameters ......: 6 2018-10-04T08:33:36: === GModelSky === 2018-10-04T08:33:36: Name ......................: Crab 2018-10-04T08:33:36: Instruments ...............: all 2018-10-04T08:33:36: Instrument scale factors ..: unity 2018-10-04T08:33:36: Observation identifiers ...: all 2018-10-04T08:33:36: Model type ................: PointSource 2018-10-04T08:33:36: Model components ..........: "PointSource" * "PowerLaw" * "Constant" 2018-10-04T08:33:36: Number of parameters ......: 6 2018-10-04T08:33:36: Number of spatial par's ...: 2 2018-10-04T08:33:36: RA .......................: 83.6331 [-360,360] deg (fixed,scale=1) 2018-10-04T08:33:36: DEC ......................: 22.0145 [-90,90] deg (fixed,scale=1) 2018-10-04T08:33:36: Number of spectral par's ..: 3 2018-10-04T08:33:36: Prefactor ................: 4.6694286848533e-15 +/- 9.16906665122574e-16 [1e-23,1e-13] ph/cm2/s/MeV (free,scale=1e-16,gradient) 2018-10-04T08:33:36: Index ....................: -2.66112738324749 +/- 0.0988449649199173 [-0,-5] (free,scale=-1,gradient) 2018-10-04T08:33:36: PivotEnergy ..............: 300000 [10000,1000000000] MeV (fixed,scale=1000000,gradient) 2018-10-04T08:33:36: Number of temporal par's ..: 1 2018-10-04T08:33:36: Normalization ............: 1 (relative value) (fixed,scale=1,gradient) 2018-10-04T08:33:36: 2018-10-04T08:33:36: +==============+ 2018-10-04T08:33:36: | Save results | 2018-10-04T08:33:36: +==============+ 2018-10-04T08:33:36: Model definition file .....: result_crab_plaw_stacked.xml 2018-10-04T08:33:36: Covariance matrix file ....: NONE 2018-10-04T08:33:36: 2018-10-04T08:33:36: Application "ctlike" terminated after 1 wall clock seconds, consuming 0.012137 seconds of CPU time.
I also added a NaN check to GCTAOnOffObservation::wstat_value so that problems can in the future be tracked down more easily. I will no merge in the code.
#2
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
- Status changed from Pull request to In Progress
- % Done changed from 100 to 50
There is still a difference in the normalisation. Here the results for the joint analysis
2018-10-08T11:45:04: Prefactor ................: 5.04442310978829e-17 +/- 3.89446356042083e-18 [1e-23,1e-13] ph/cm2/s/MeV (free,scale=1e-16,gradient) 2018-10-08T11:45:04: Index ....................: -2.63791542087657 +/- 0.0845343261931618 [-0,-5] (free,scale=-1,gradient) 2018-10-08T11:45:04: PivotEnergy ..............: 1000000 [10000,1000000000] MeV (fixed,scale=1000000,gradient)
and here for the stacked analysis
2018-10-08T13:01:40: Prefactor ................: 2.05784483532498e-16 +/- 1.63719727869171e-17 [1e-23,1e-13] ph/cm2/s/MeV (free,scale=1e-16,gradient) 2018-10-08T13:01:40: Index ....................: -2.63551070265515 +/- 0.0862634071633871 [-0,-5] (free,scale=-1,gradient) 2018-10-08T13:01:40: PivotEnergy ..............: 1000000 [10000,1000000000] MeV (fixed,scale=1000000,gradient)
#3
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
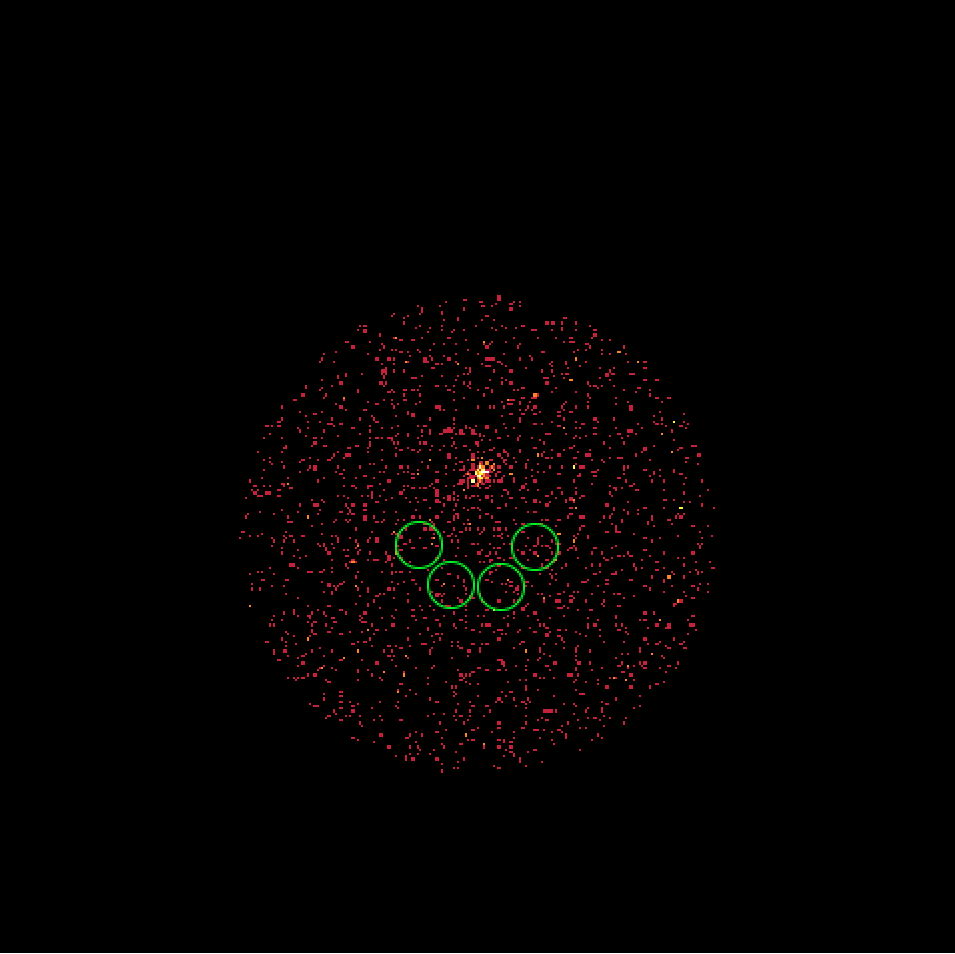
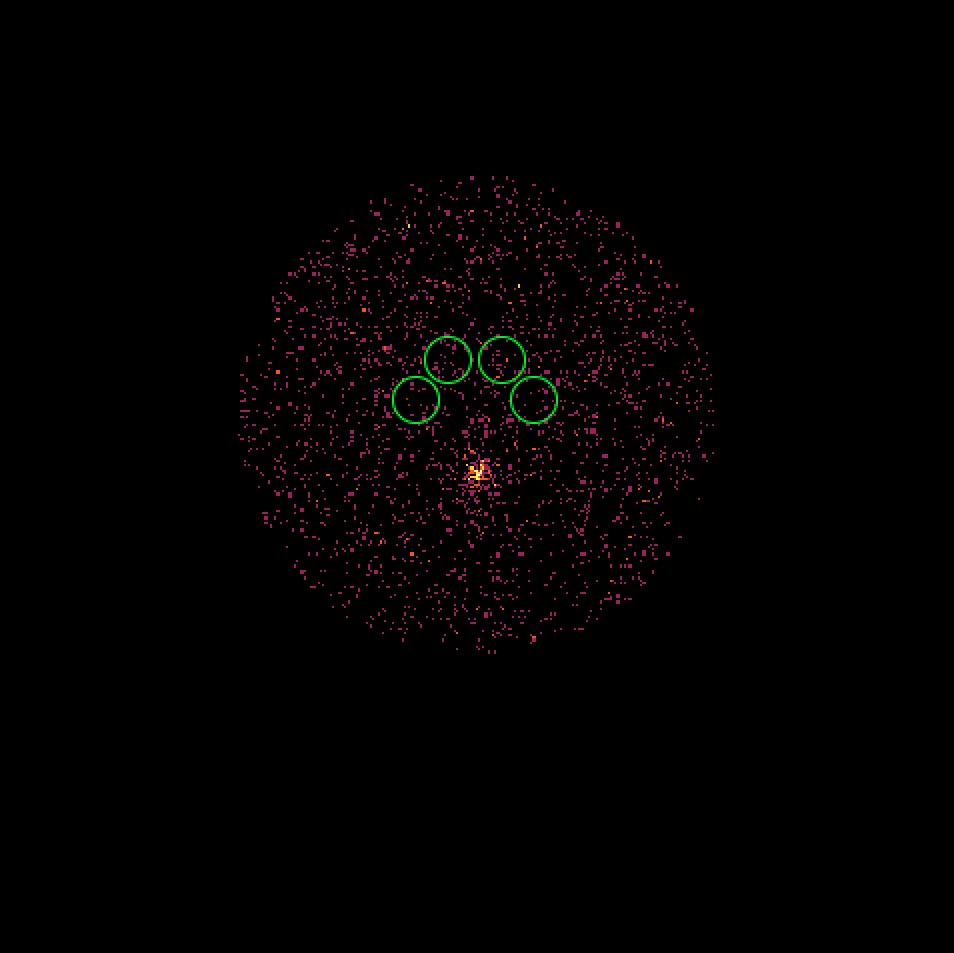
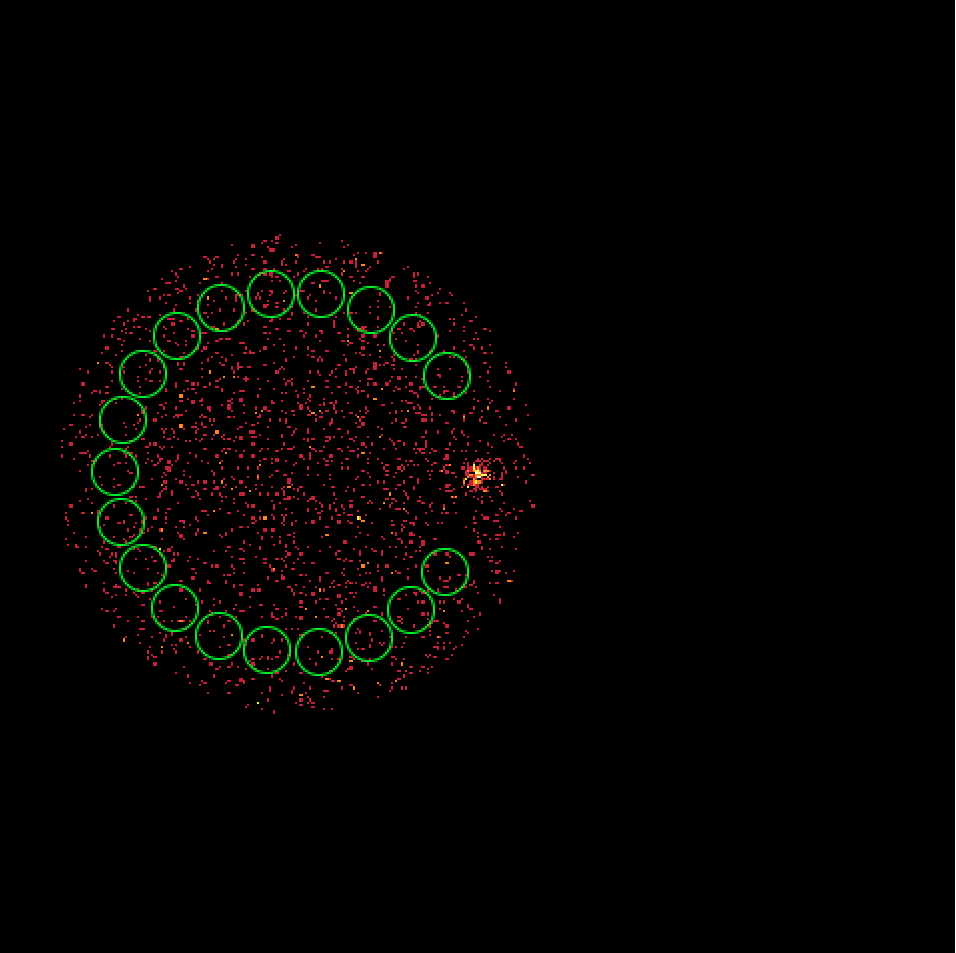
- File skymap_023523.png added
- File skymap_023526.png added
- File skymap_023559.png added
- File skymap_023592.png added
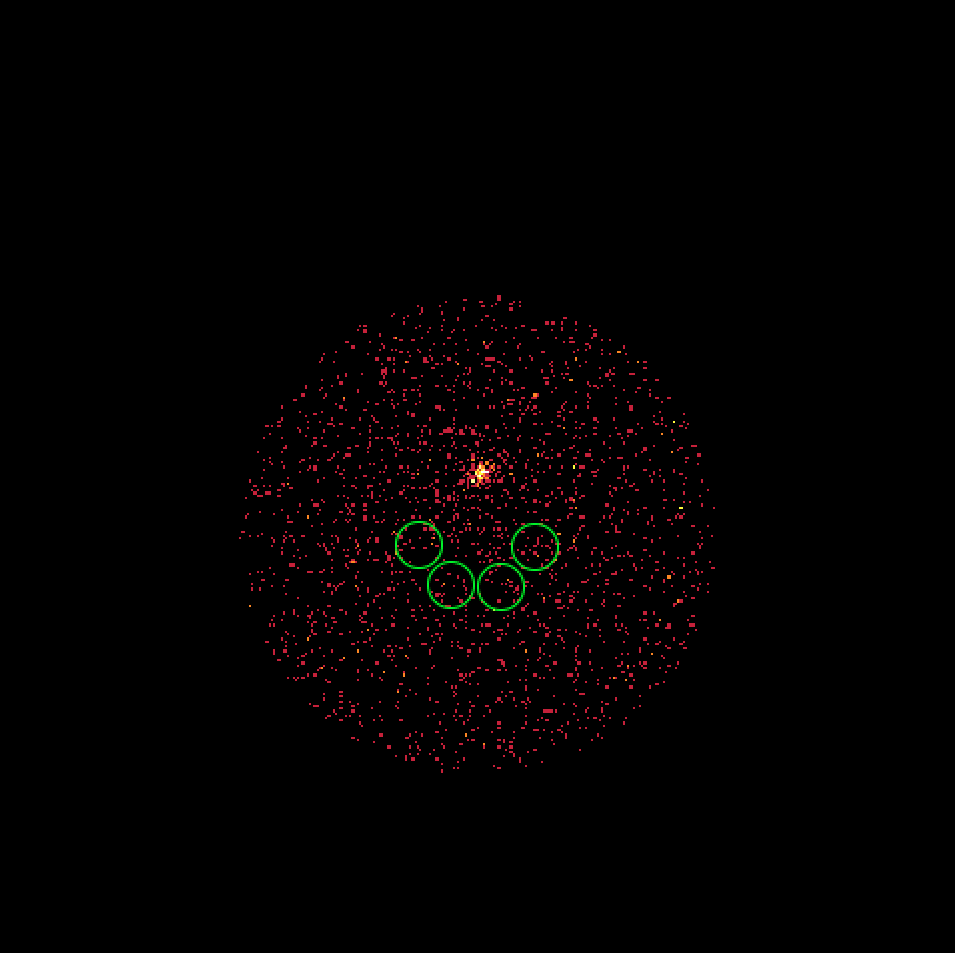
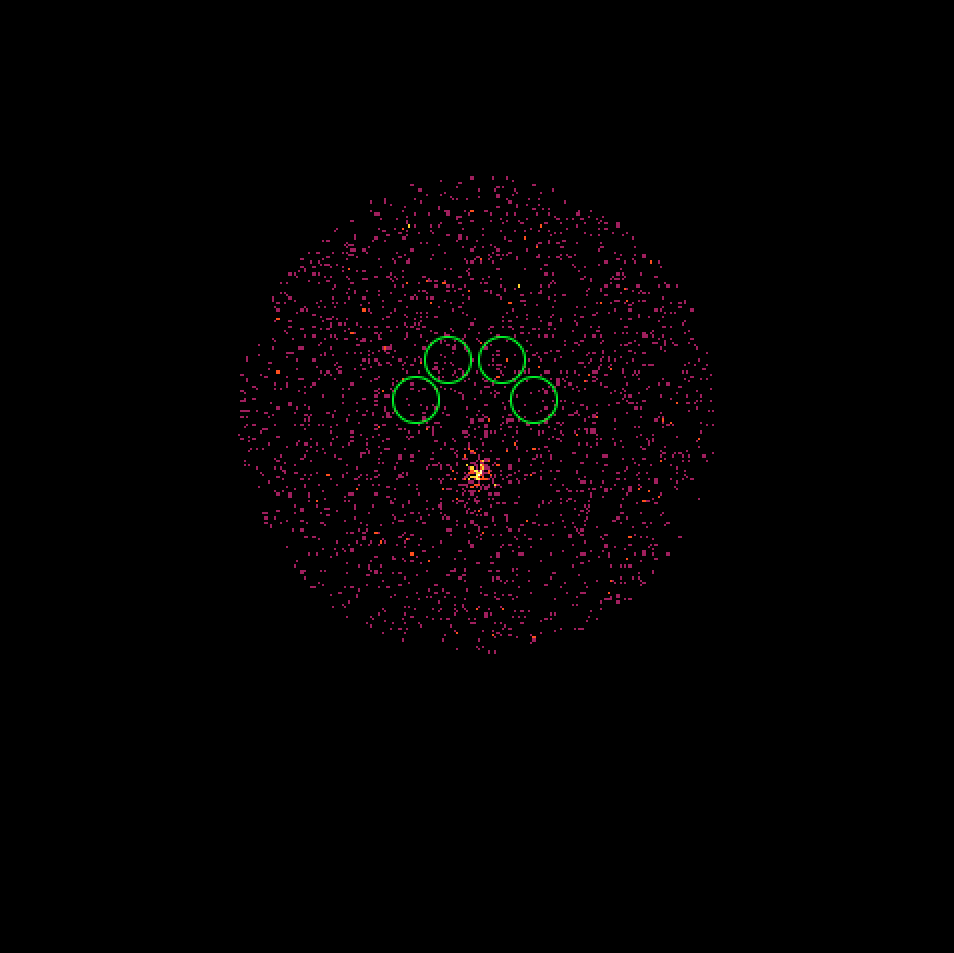
It turns out that the 4 H.E.S.S. runs combine observations with a different offset angle of the Crab and hence the background rate of the Off regions vary between the first two and the second two runs. We always need an assumption about the spatial background rate distribution, and in absence of a template the probably best thing to do is to assume that the background rate is constant. That’s what the actual code does. This assumption is however violated in the present case, and I see no obvious solution to this problem. Stacking seems simply not possible in the absence of a background template!
 |
 |
 |
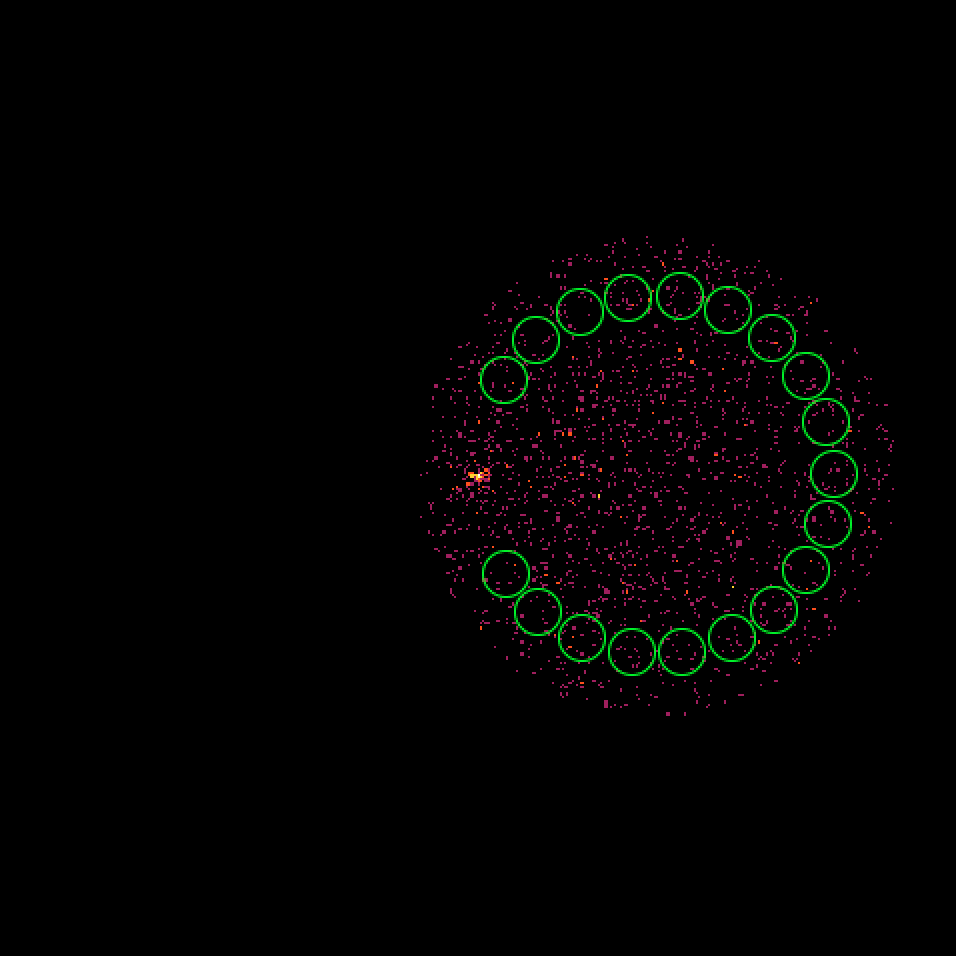 |
#4
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
I tried to group the runs, making a stacked On/Off observation of the first two and the second two runs, and then fit the runs jointly. Still, the result is different to the joint fitting result:
2018-10-08T13:10:25: Prefactor ................: 1.01320579621174e-16 +/- 8.17283310635949e-18 [1e-23,1e-13] ph/cm2/s/MeV (free,scale=1e-16,gradient) 2018-10-08T13:10:25: Index ....................: -2.64470055097098 +/- 0.0879902336299644 [-0,-5] (free,scale=-1,gradient) 2018-10-08T13:10:25: PivotEnergy ..............: 1000000 [10000,1000000000] MeV (fixed,scale=1000000,gradient)
#5
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
- % Done changed from 50 to 60
It turned out that the exposure time was not correctly written by the GPha write method (it was in fact overwritten by the exposure time of the first stacked run). I corrected this bug and now the results look more reasonable:
2018-10-08T13:50:18: Prefactor ................: 5.07679333256999e-17 +/- 4.09478853758408e-18 [1e-23,1e-13] ph/cm2/s/MeV (free,scale=1e-16,gradient) 2018-10-08T13:50:18: Index ....................: -2.64578609753126 +/- 0.0880065744992247 [-0,-5] (free,scale=-1,gradient) 2018-10-08T13:50:18: PivotEnergy ..............: 1000000 [10000,1000000000] MeV (fixed,scale=1000000,gradient)
#6
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
Here now the result for the stacked analysis of all 4 runs.
2018-10-08T13:57:42: Prefactor ................: 5.15734295281727e-17 +/- 4.10298669503984e-18 [1e-23,1e-13] ph/cm2/s/MeV (free,scale=1e-16,gradient) 2018-10-08T13:57:42: Index ....................: -2.63605170766175 +/- 0.0862709625106715 [-0,-5] (free,scale=-1,gradient) 2018-10-08T13:57:42: PivotEnergy ..............: 1000000 [10000,1000000000] MeV (fixed,scale=1000000,gradient)
They are not too bad, given the simplistic hypothesis about the background rate.
#7
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
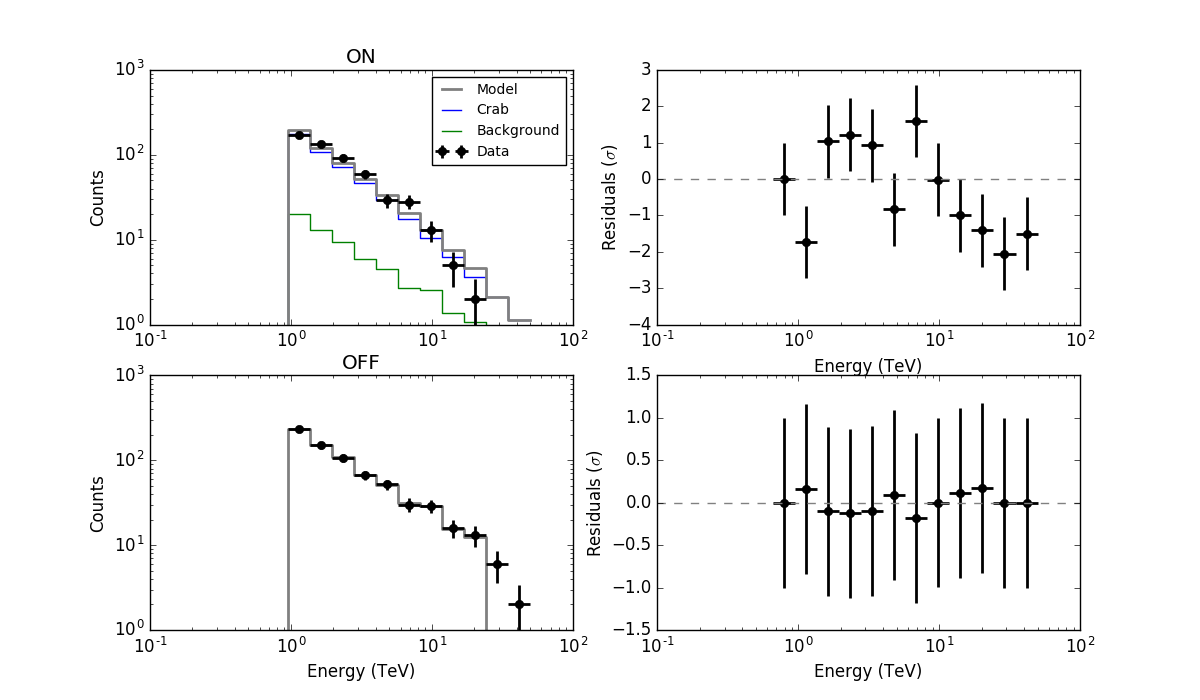
- File residuals.png added
Concerning csresspec, the issue is that you need to run the script using the statistic=wstat option:
$ csresspec statistic=wstat components=yes Input event list, counts cube, or observation definition XML file [obs_crab_onoff_stacked.xml] Input model definition XML file [results.xml] Residuals computation algorithm (SUB|SUBDIV|SUBDIVSQRT|SIGNIFICANCE) [SIGNIFICANCE] Output residual spectrum file [resspec.fits] $CTOOLS/share/examples/python/show_residuals.py resspec.fits

#8
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
- Duplicated by Bug #2696: stacked on/off analysis added
#9
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
- Status changed from In Progress to Feedback
- % Done changed from 60 to 90
Merged into devel. Luigi, Andreas, can you tell me how this fix works on your side?
#10
 Updated by Specovius Andreas over 6 years ago
Updated by Specovius Andreas over 6 years ago
Knödlseder Jürgen wrote:
Merged into
devel. Luigi, Andreas, can you tell me how this fix works on your side?
Your fix for alpha seems to work fine for me.
I also saw the discrepancies between stacked and joint analysis you mentioned above. For msh1552 (HESS PDR) the normalization stays roughly the same while the index jumps from 2.2 (stacked) to 2.4 (joint). This might be an effect of the lower statistics per run compared to pks or crab ... (?!)
I also noticed that effective area below threshold is zero and energy dispersion for Etrue<Ethres redistributing photons to Ereco>Ethres is zero, both resulting in zero probability of near but below threshold photons to contribute to the predicted flux (we had this recently for the joint case). I am not sure whether we already talked about this but is this another issue?
#11
 Updated by Tibaldo Luigi over 6 years ago
Updated by Tibaldo Luigi over 6 years ago
I will test the new devel code tomorrow, in the mean time two remarks.
1) For stacking multiple runs without a background template I recalled what is done in HESS. The OFF counts themselves are used as a proxy of the background response of each run, so that
BACKSCAL = Sum_i alpha_i * NOFF_i / Sum_i NOFF_i
I think it would be better to implement this formula for the case without background template, rather than assuming the background rate is the same for all runs. Does that make sense to you?
2) I was already using statistic=WSTAT for csresspec, so that was definitely not the cause of the problem I reported. I will tests again tomorrow to see if the problem is still present in the latest version.
#12
 Updated by Tibaldo Luigi over 6 years ago
Updated by Tibaldo Luigi over 6 years ago
- File joint_stacked.png added
1) With the current devel version I obtain comparable results for joint and stacked analysis of the Crab. I still recommend to implement the weighting of BACKSCAL on N_OFF for stacking without background template. This will allow for more general cases to be accommodated. Josh told me that the same strategy is used in VERITAS as well.
2) I still have problems with calculating stacked residuals from multiple On/Off observations that were analyzed jointly. I’m copying here my code and the results
observations = 'obs_crab_selected.xml' models = 'crab_models.xml' onoff_obs = 'obs_crab_onoff_joint.xml' phagen = cscripts.csphagen() phagen['inobs'] = observations phagen['inmodel'] = models phagen['srcname'] = 'Crab' phagen['outobs'] = onoff_obs phagen['ebinalg'] = 'LOG' phagen['emin'] = 0.66 phagen['emax'] = 30. phagen['enumbins'] = 20 phagen['coordsys'] = 'CEL' phagen['ra'] = 83.63 phagen['dec'] = 22.01 phagen['rad'] = 0.2 phagen['bkgmethod'] = 'REFLECTED' phagen['use_irf_bkg'] = False phagen['maxoffset'] = 2.0 phagen['stack'] = False phagen.execute() fit_models = 'crab_fit_models_joint.xml' like = ctools.ctlike() like['statistic'] = 'WSTAT' like['inobs'] = onoff_obs like['inmodel'] = models like['outmodel'] = fit_models like.execute() residuals = 'residuals_joint_stacked.fits' res = cscripts.csresspec() res['statistic'] = 'WSTAT' res['inobs'] = onoff_obs res['inmodel'] = fit_models res['components'] = True res['algorithm'] = 'SIGNIFICANCE' res['outfile'] = residuals res['stack'] = True res.execute()
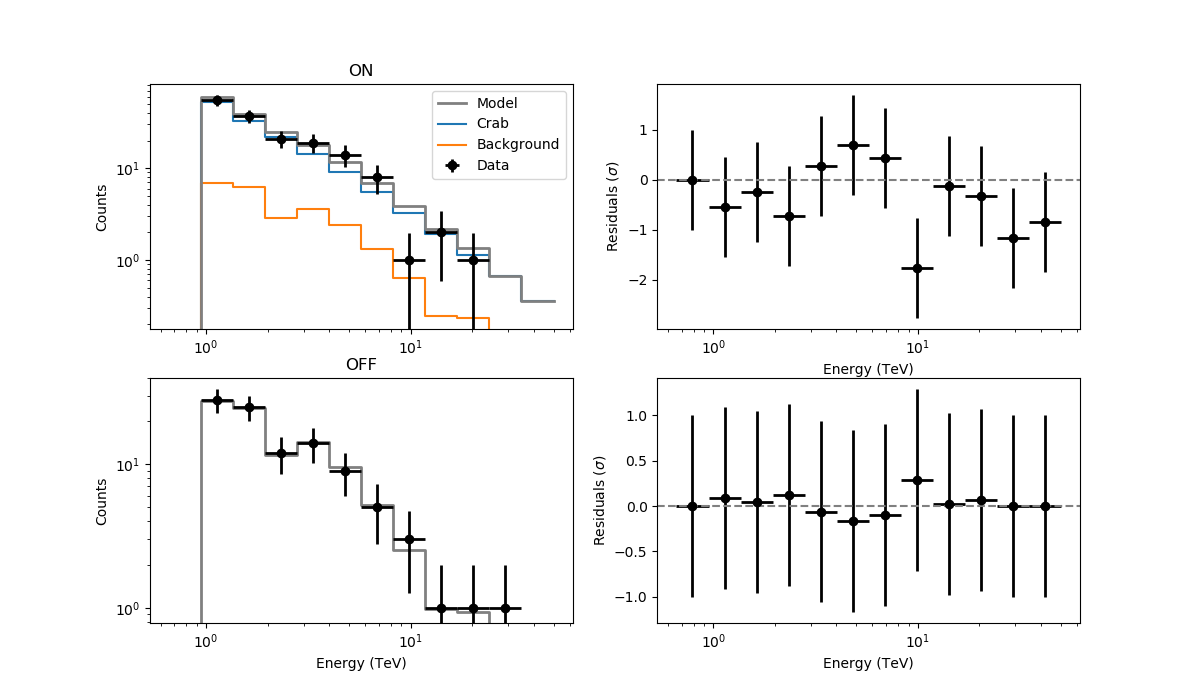
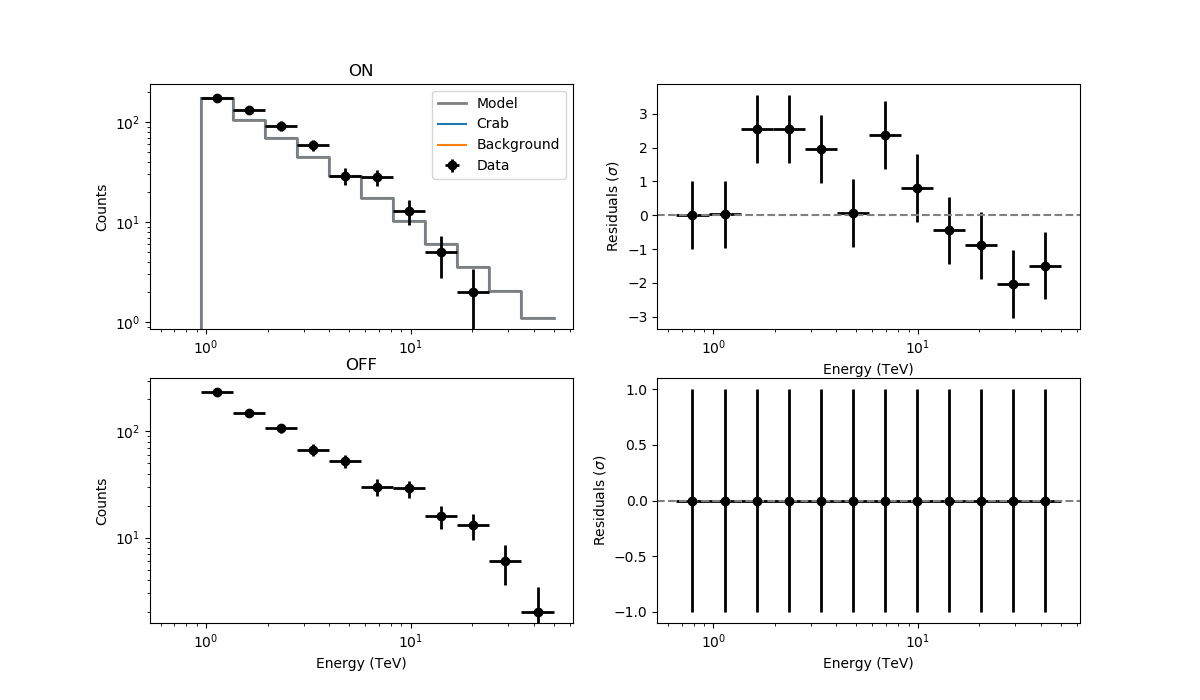
The resulting residuals are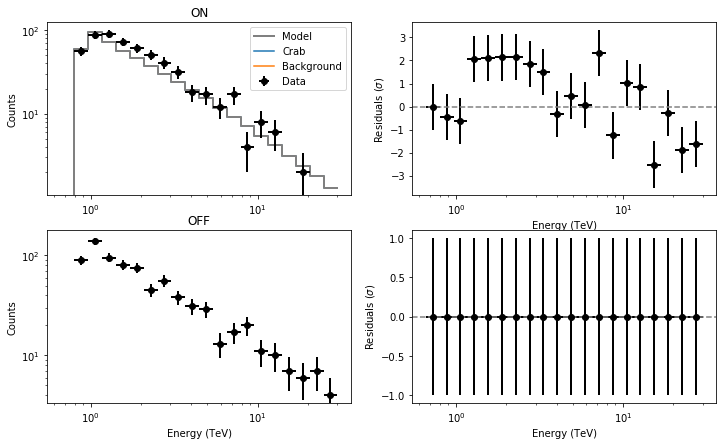
I’m using WSTAT in csresspec. The background model is identically 0. Please, let me know if there are any issues, but otherwise could it be something in csresspec that does not work properly without a background template?
#13
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
- Status changed from Feedback to In Progress
Just to recap where we are for the 9th coding sprint:
The GCTAOnOffObservation::compute_bgd computes the BACKRESP column in the Off spectrum, which without a background model is just the solid angle of the Off region region for a given observations. Note that in the commit before, the values of the BACKRESP column were simply set to 0 in case that no background model exists.
The GCTAOnOffObservation::compute_alpha computes the alpha factor, which without a background model is just the energy independent ratio between the solid angle of the On and Off regions.
The wstat method is not using the BACKRESP column information, it is however used when stacking in the GCTAOnOffObservation::GCTAOnOffObservation(const GObservations& obs) constructor:
// Compute background scaling factor contribution
for (int i = 0; i < m_on_spec.size(); ++i) {
double background = onoff->off_spec()["BACKRESP"][i];
double alpha = onoff->on_spec().backscal(i);
double scale = alpha * background * exposure;
m_on_spec.backscal(i, m_on_spec.backscal(i) + scale);
}
According to Luigi’s proposal, in case that background is zero, one should probably replace the code as follows (two places):
// Compute background scaling factor contribution
for (int i = 0; i < m_on_spec.size(); ++i) {
double background = onoff->off_spec()["BACKRESP"][i];
double alpha = onoff->on_spec().backscal(i);
double scale = 0.0;
if (background > 0.0) {
scale = alpha * background * exposure;
}
else {
scale = alpha * onoff->off_spec()[i];
}
m_on_spec.backscal(i, m_on_spec.backscal(i) + scale);
}
and
// Compute stacked background scaling factor
for (int i = 0; i < m_on_spec.size(); ++i) {
double norm = m_off_spec["BACKRESP"][i];
if (norm > 0.0) {
double scale = m_on_spec.backscal(i) / norm;
m_on_spec.backscal(i,scale);
}
else {
double scale = m_on_spec.backscal(i) / m_off_spec[i];
m_on_spec.backscal(i,scale);
}
}
at the end. in
GCTAOnOffObservation::compute_bgd, the BACKRESP column should be set to zero by removing the else section after // ... otherwise
else {
// Loop over regions
for (int k = 0; k < off.size(); ++k) {
Hope this does the job. The change should probably be implemented using a compile flag so that one can easily switch forth and back between the code versions.
#14
 Updated by Knödlseder Jürgen about 6 years ago
Updated by Knödlseder Jürgen about 6 years ago
I this one fixed now?
#15
 Updated by Tibaldo Luigi about 6 years ago
Updated by Tibaldo Luigi about 6 years ago
Yes, based on the latest tests I did everything works fine!
#16
 Updated by Knödlseder Jürgen about 6 years ago
Updated by Knödlseder Jürgen about 6 years ago
- Status changed from In Progress to Closed
- % Done changed from 90 to 100