Change request #2685
Improve handling of varying energy thresholds in stacked analysis
| Status: | Closed | Start date: | 09/21/2018 | |
|---|---|---|---|---|
| Priority: | Normal | Due date: | ||
| Assigned To: | % Done: | 100% | ||
| Category: | - | |||
| Target version: | 1.6.0 | |||
| Duration: |
Description
When analysing residual spectra of the Crab for the H.E.S.S. data release the following run
$ csresspec debug=yes edisp=yes components=yes logfile=resspec_on.log Input event list, counts cube, or observation definition XML file [obs_crab_selected.xml] Algorithm for defining energy bins (FILE|LIN|LOG) [LOG] Start value for first energy bin in TeV [0.66] Stop value for last energy bin in TeV [100.0] Number of energy bins (1-200) [40] Stack observations? [yes] Coordinate System (CEL|GAL) [CEL] Projection method (AIT|AZP|CAR|GLS|MER|MOL|SFL|SIN|STG|TAN) [CAR] First coordinate of image/source region center in degrees (RA or galactic l) (0-360) [83.63] Second coordinate of image/source region center in degrees (DEC or galactic b) (-90-90) [22.01] Size of the X axis in pixels [200] Size of the Y axis in pixels [200] Pixel size (deg/pixel) [0.02] Input model definition XML file [crab_results.xml] Mask data to calculate residuals in ROI? [yes] RA for ROI centre (degrees) (0-360) [83.63] Dec for ROI centre (degrees) (-90-90) [22.01] Radius of ROI (degrees) (0-180) [0.2] Input exclusion region file in ds9 format [NONE] Residuals computation algorithm (SUB|SUBDIV|SUBDIVSQRT|SIGNIFICANCE) [SIGNIFICANCE] Output residual spectrum file [resspec_on.fits]
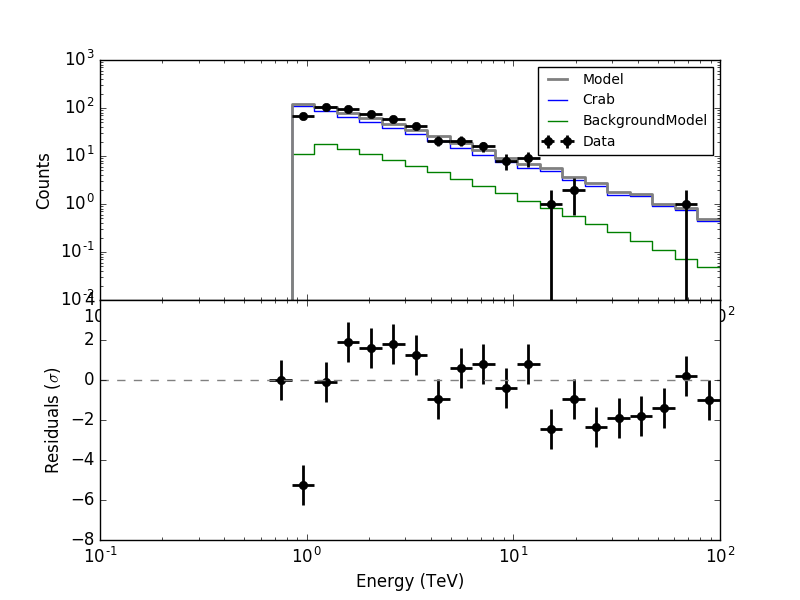
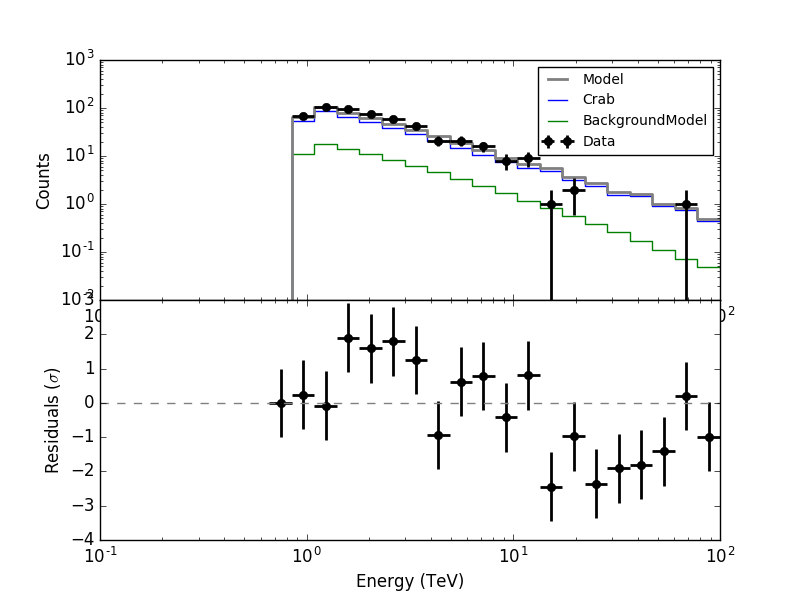
led to the following residual spectrum:
 For the second energy bin, running from 0.848 - 1.090 TeV, the Crab model value is well above the measured number of counts. The reason is the following.
For the second energy bin, running from 0.848 - 1.090 TeV, the Crab model value is well above the measured number of counts. The reason is the following. ctbin only fills counts cube bins if they are fully covered by the energy range of the events. The energy thresholds of the four runs being 0.871 TeV, 0.708 TeV, 0.661 TeV and 0.871 TeV the filling of the counts cube bins is as follows:
- Bin 0 (0.66 - 0.848 TeV): no runs
- Bin 1 (0.848 - 1.0904 TeV): runs 2 & 3
- Bin 2 (1.0904 - 1.401 TeV): all runs
The effective area is however for a finer energy binning. Effective area is also given in true energies. All four runs contribute to the effective area of bin 1, since the energy nodes of the effective area matching bin 1 are all above the highest threshold.
Now the counts cube weight as computed by ctbin is 1 for the bin 1 at the location of the Crab, hence the Crab model is computed as if all 4 runs were contributing to data. It needs to be checked how the computation should be done correctly, and the code needs to be modified.
Recurrence
No recurrence.
History
#1
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
- File resspec_on_corrected.png added
- Status changed from New to In Progress
- Assigned To set to Knödlseder Jürgen
- Target version set to 1.6.0
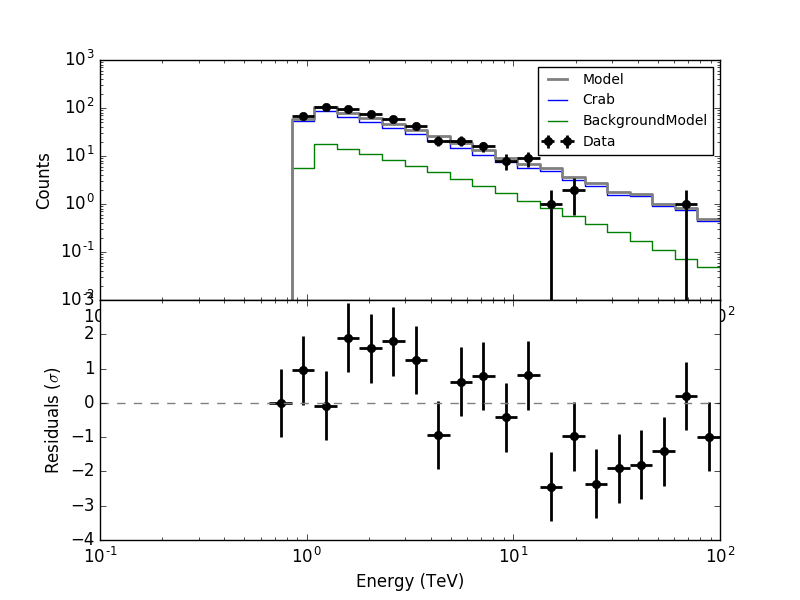
I added some code to ctbin so that the actual weight of a given pixel is computed by the ratio of the runs that are used divided by the total number of runs. Specifically, in ctbin::set_weights the code is now
// Loop over all energy layers of counts cube
for (int iebin = 0; iebin < m_ebounds.size(); ++iebin){
// Add livetime to weight sum before
#pragma omp critical(ctbin_set_weights)
m_weights_sum(pixel, iebin) += livetime;
// Skip energy layer if the usage flag is false
if (!usage[iebin]) {
continue;
}
// Add livetime to weight
#pragma omp critical(ctbin_set_weights)
m_weights(pixel, iebin) += livetime;
where the weight of a bin is computed from the livetime (so the actual weight value is the ratio of use livetime with respect to total livetime of all runs for that pixel).
The normalization is then done in ctbin::run:
// Normalize weight array by weight sum
for (int pixel = 0; pixel < m_counts.npix(); ++pixel) {
for (int iebin = 0; iebin < m_ebounds.size(); ++iebin){
if (m_weights_sum(pixel, iebin) > 0.0) {
m_weights(pixel, iebin) /= m_weights_sum(pixel, iebin);
}
}
}
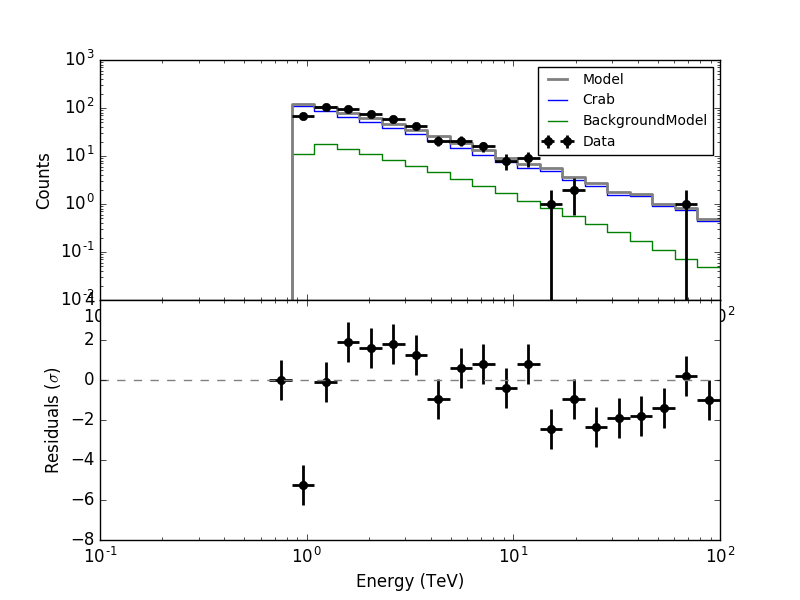
It looks like the residuals are now okay:
#2
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
- % Done changed from 0 to 50
#3
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
- File resspec_5nodes.png added
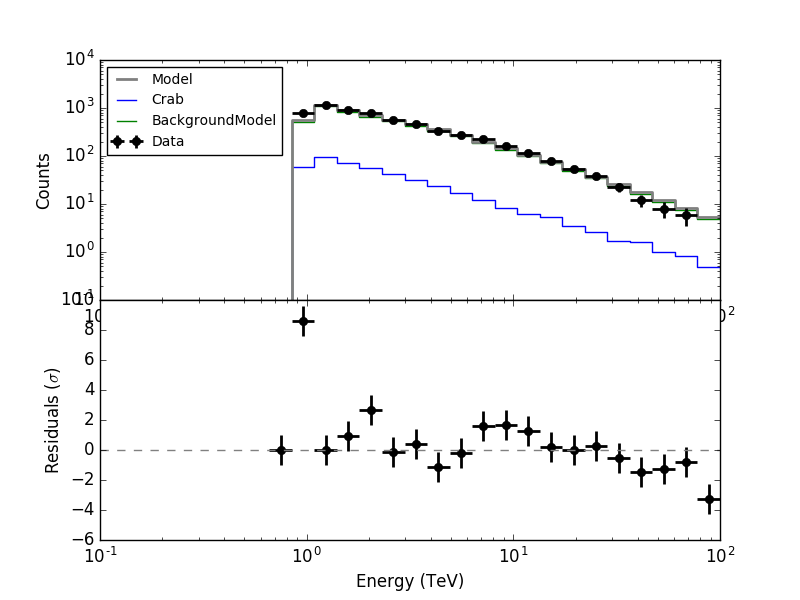
However when the integration is done over the full ROI, there is now problem: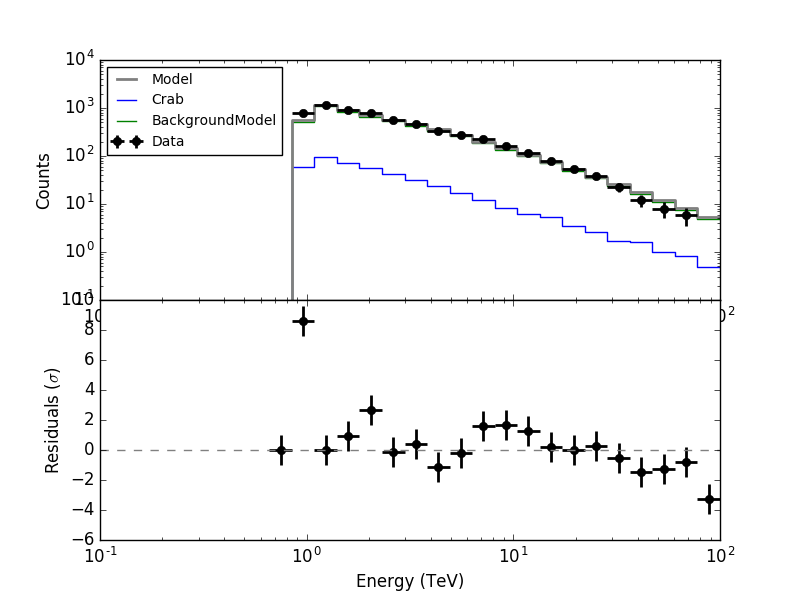
The issue is that the new weighting scheme is appropriate for the signal that is convolved with the IRF, but not for the background that is not convolved with the IRF.
#4
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
This looks like a fundamental issue. In GObservation::likelihood_poisson_binned there is the following code:
// Get model and derivative
double model = this->model(models, *bin, &wrk_grad);
// Multiply model by bin size
model *= bin->size();
The bin weighting is done in the
size() method. The model variable combines IRF and background models, hence there is for the moment no other option that “unweighting” the background model so that weighting recovers the original value.#5
 Updated by Knödlseder Jürgen over 6 years ago
Updated by Knödlseder Jürgen over 6 years ago
- File resspec_new1.png added
- File resspec_on_new1.png added
- % Done changed from 50 to 80
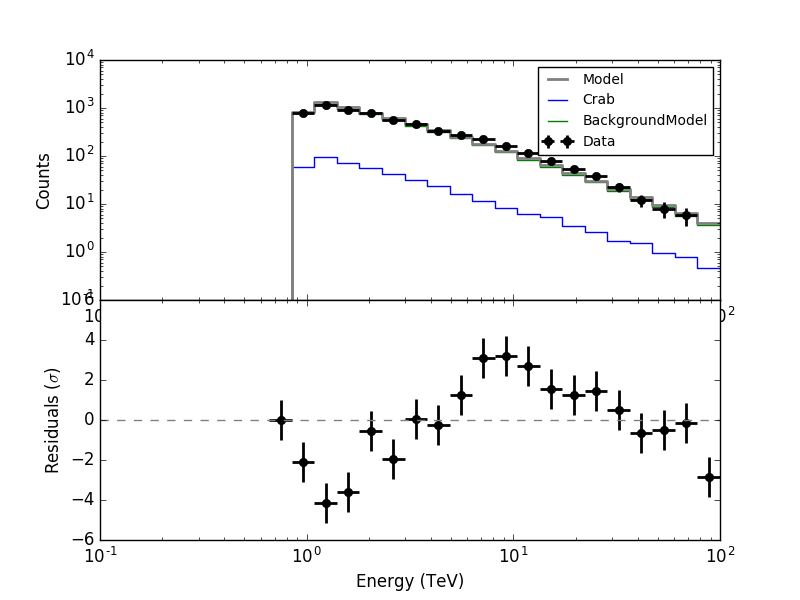
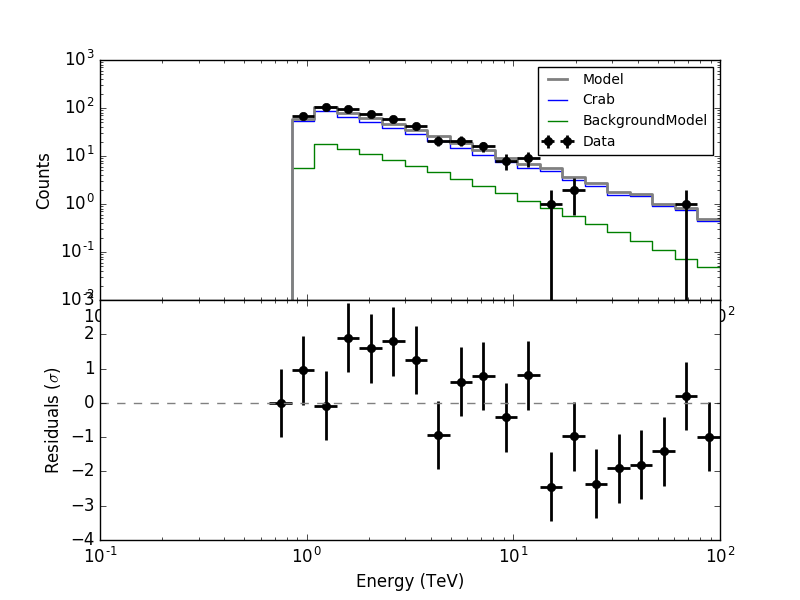
I made all the necessary changes. Now in ctbkgcube the background cube response is unweighted before storing. To make all things work I needed to modify also obsutils so that a counts cube is passed to ctbkgcube in the response generation.
Here are now the residuals for the full data space (left) and the On region around the Crab (right). In both cases there are no longer threshold effects visible.
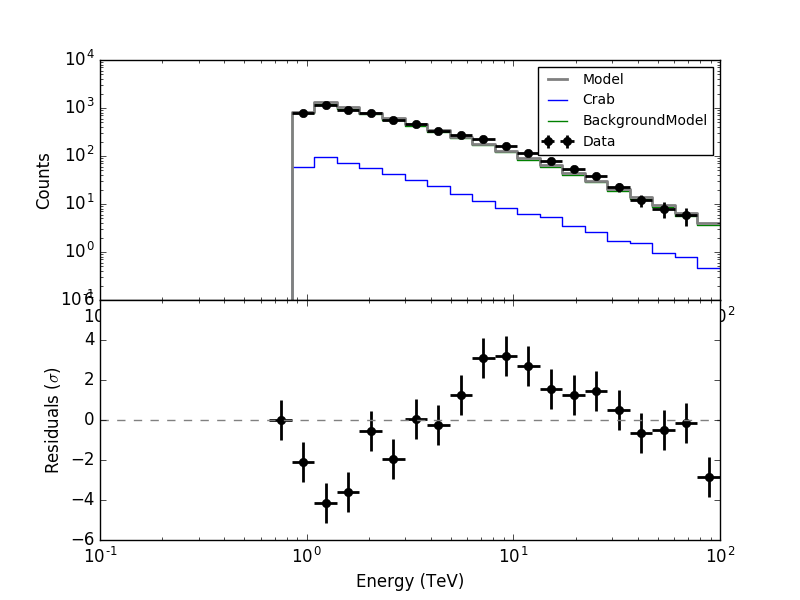 |
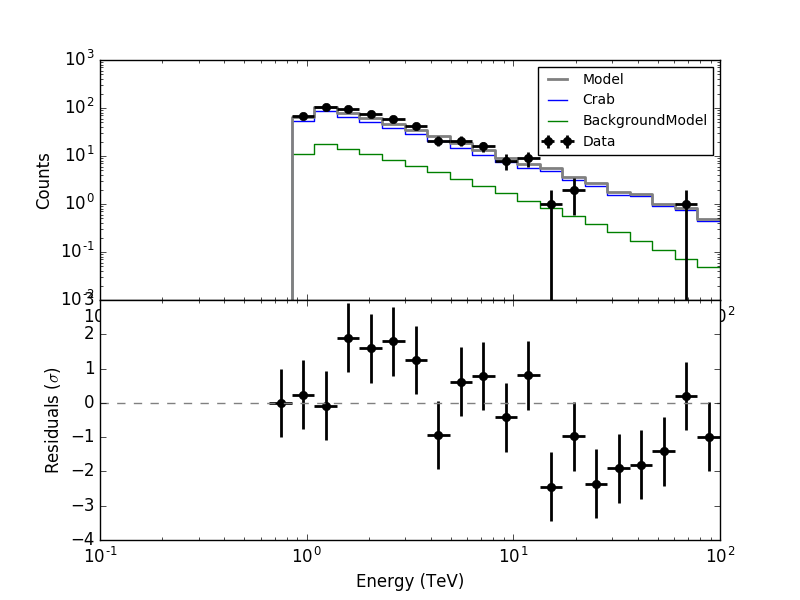 |
#6
 Updated by Knödlseder Jürgen about 6 years ago
Updated by Knödlseder Jürgen about 6 years ago
- Status changed from In Progress to Closed
- % Done changed from 80 to 100