Bug #1503
Pull distributions of stacked analysis look shifted
| Status: | Closed | Start date: | 07/03/2015 | |
|---|---|---|---|---|
| Priority: | Normal | Due date: | ||
| Assigned To: | % Done: | 100% | ||
| Category: | - | |||
| Target version: | 1.0.0 | |||
| Duration: |
Description
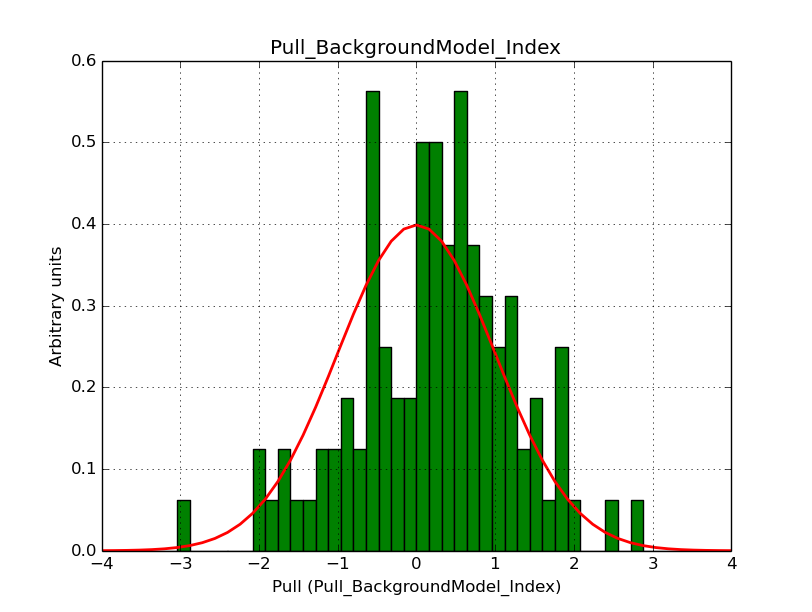
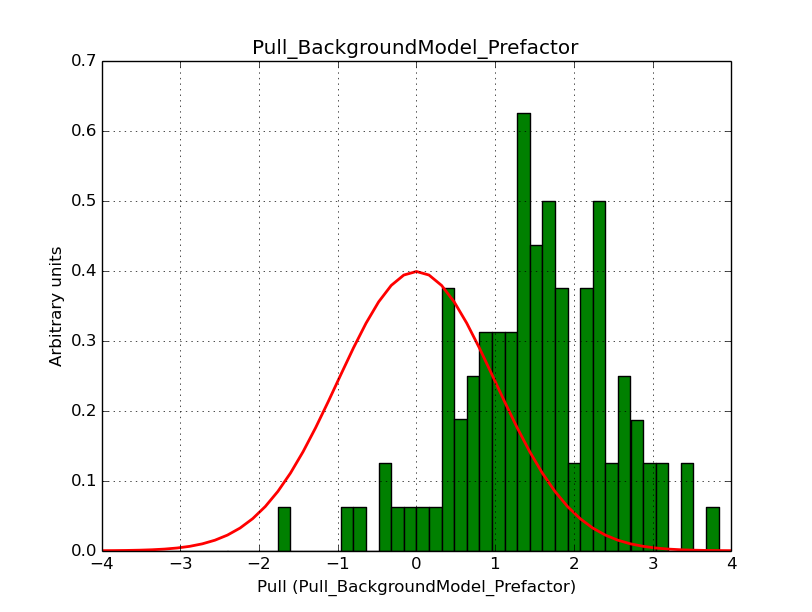
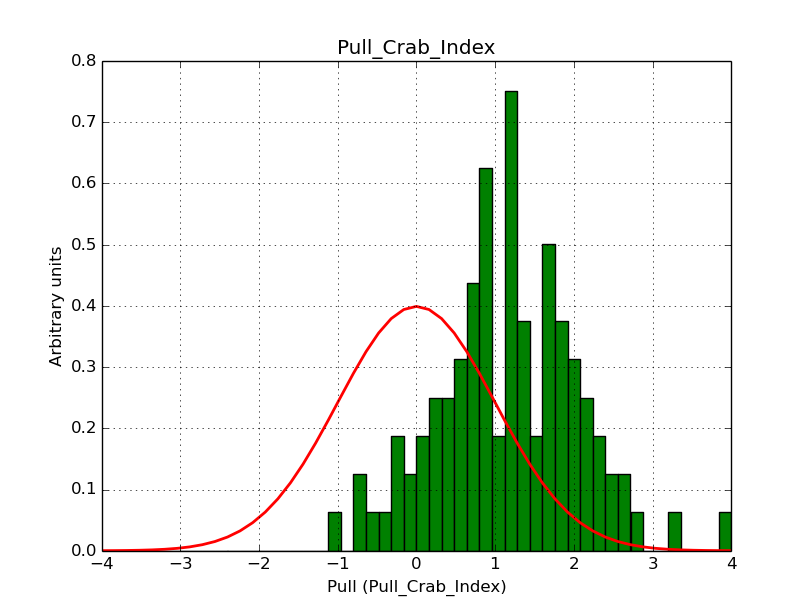
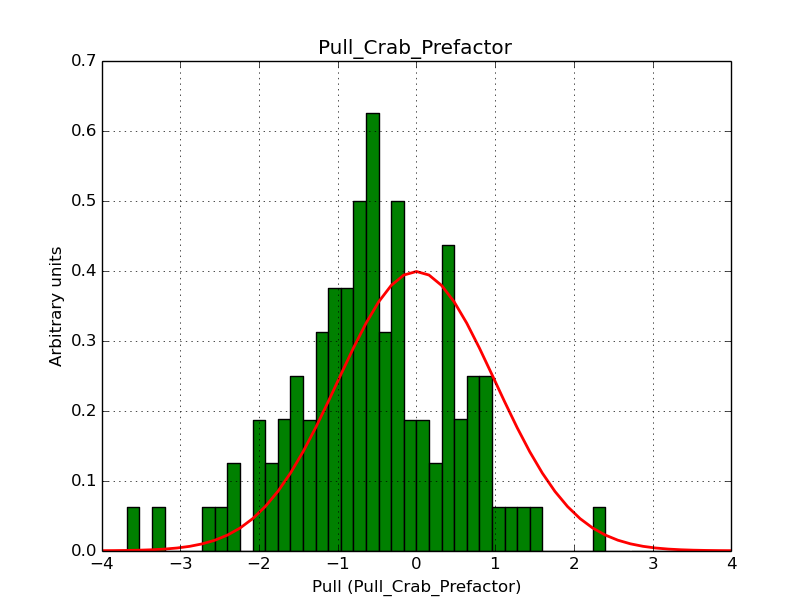
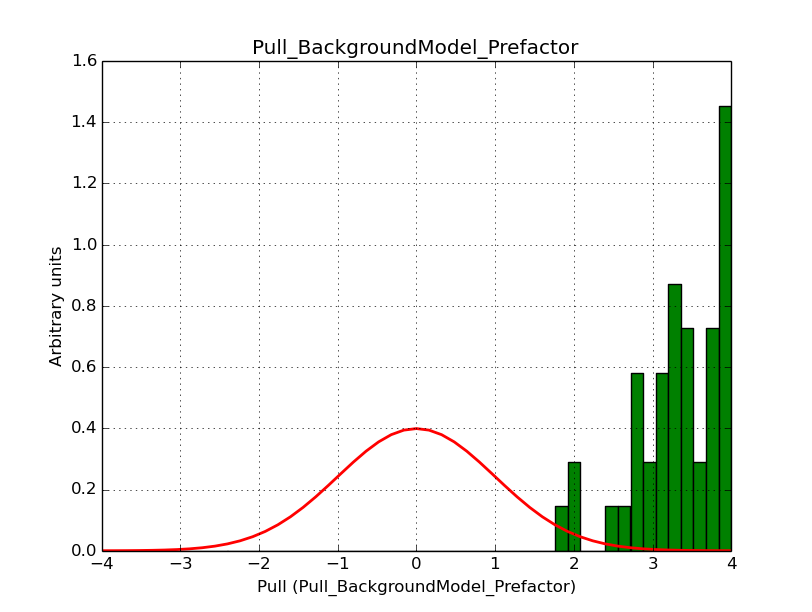
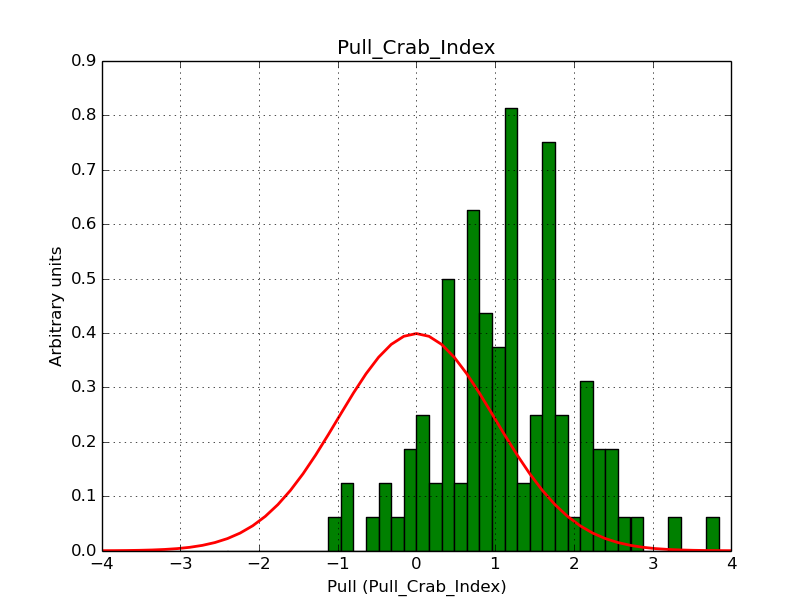
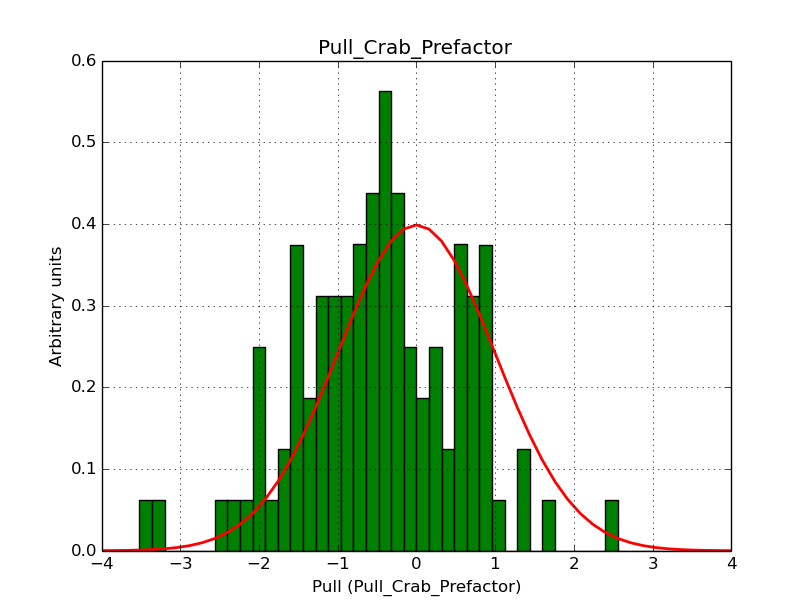
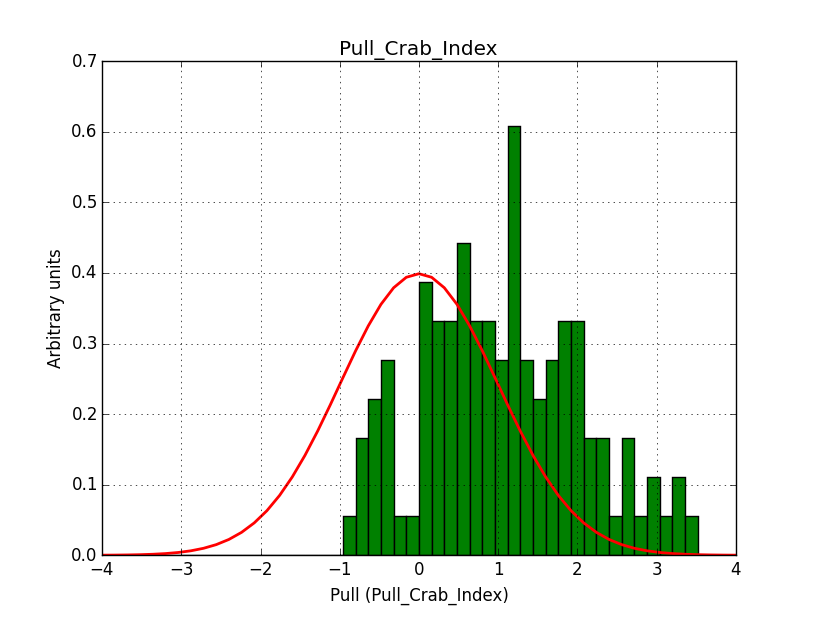
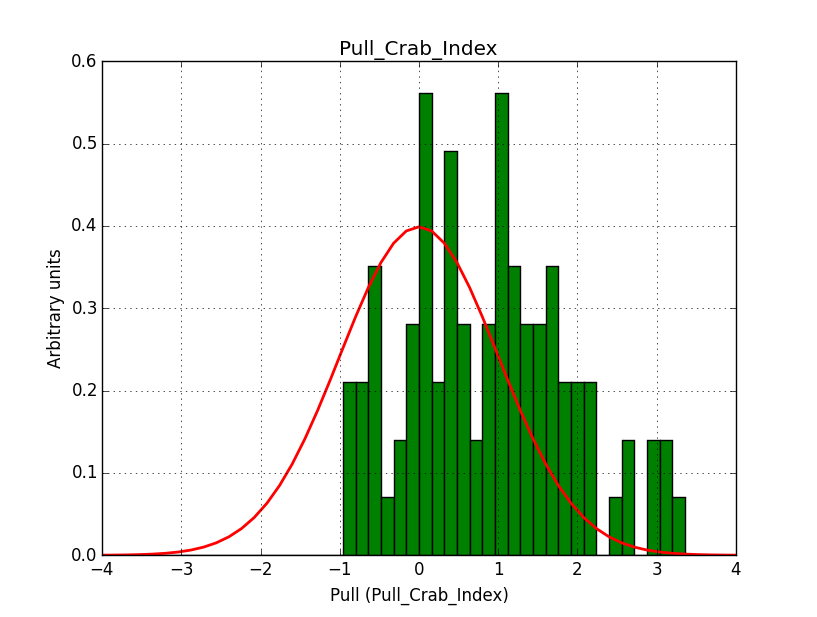
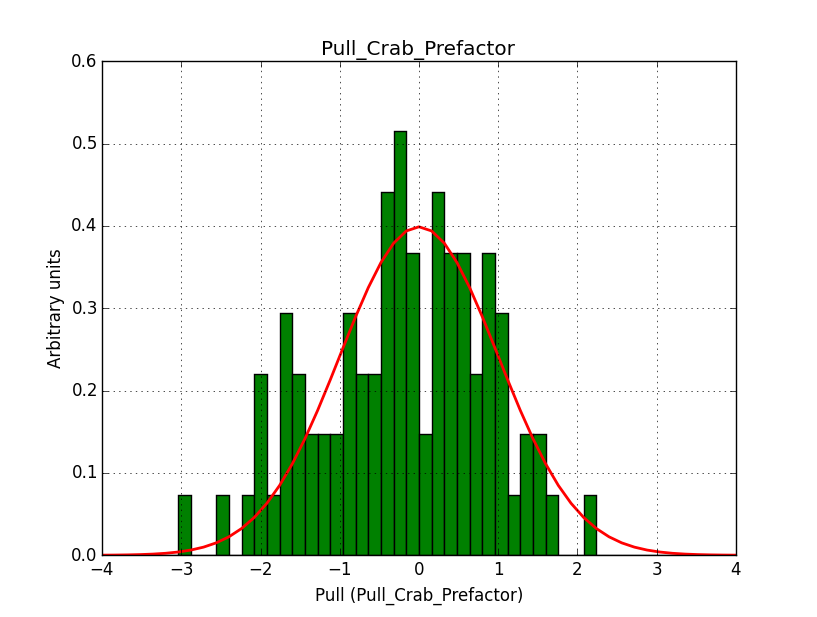
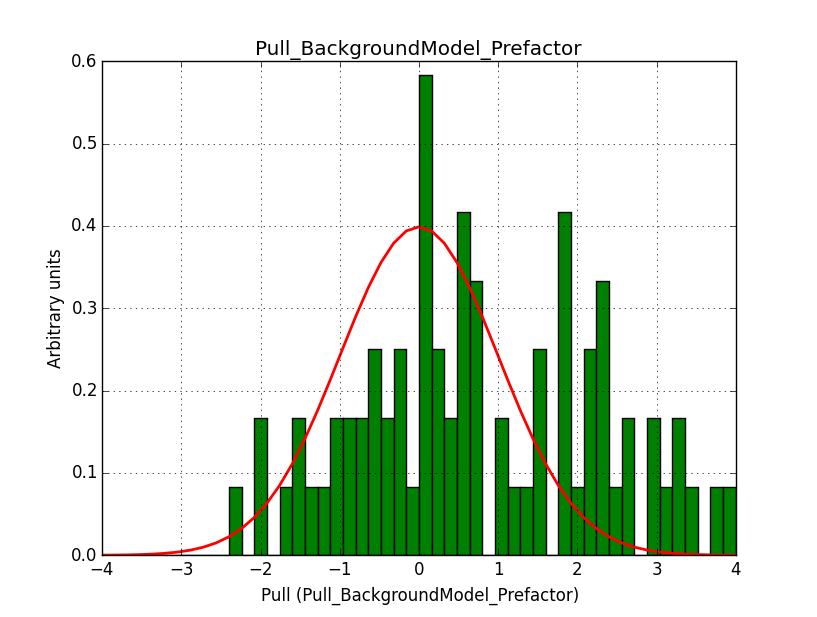
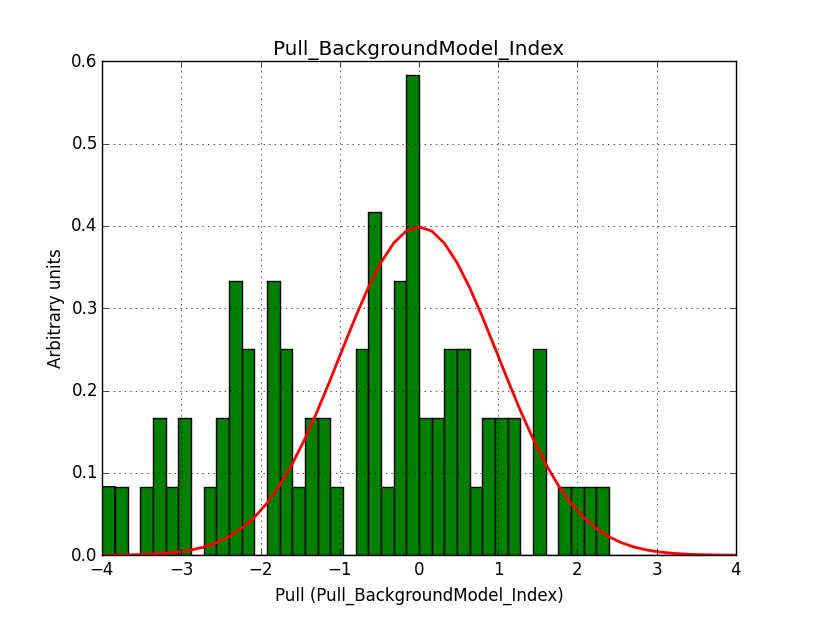
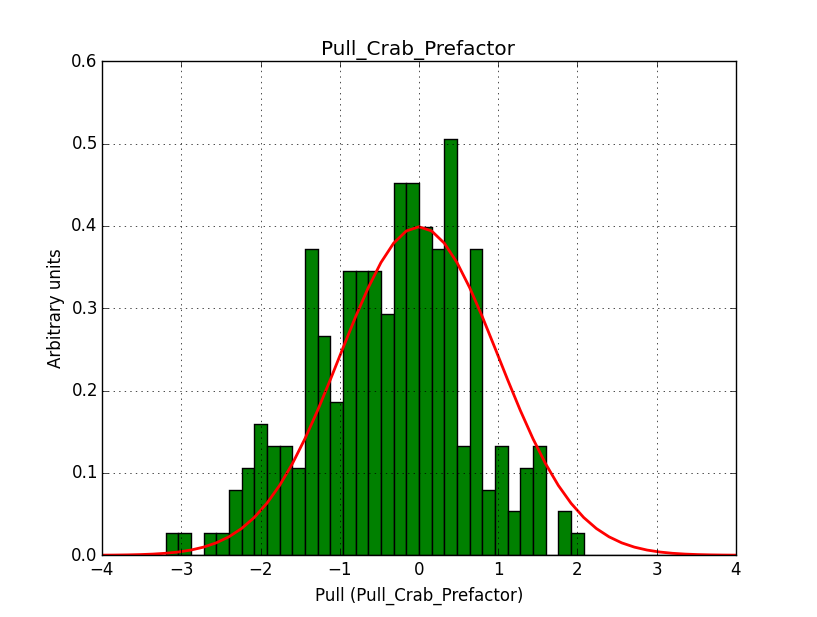
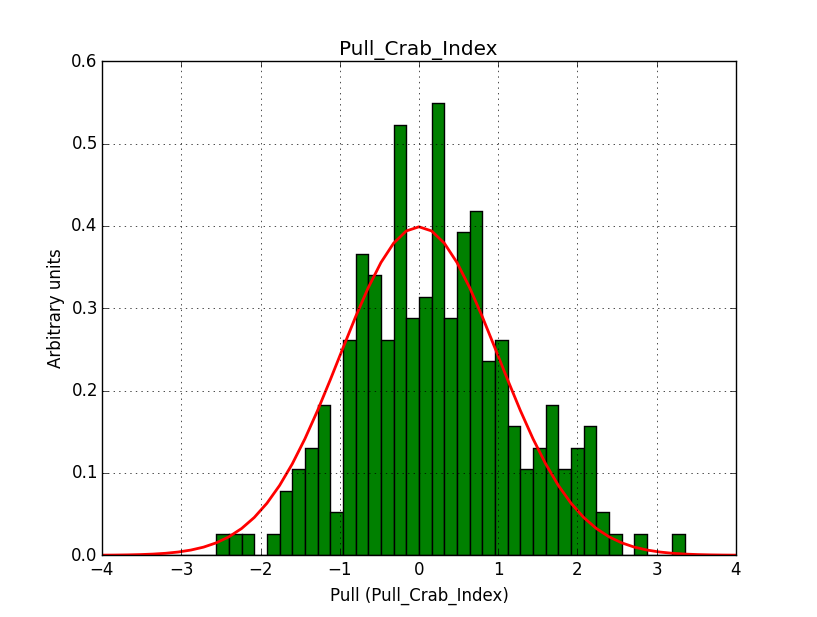
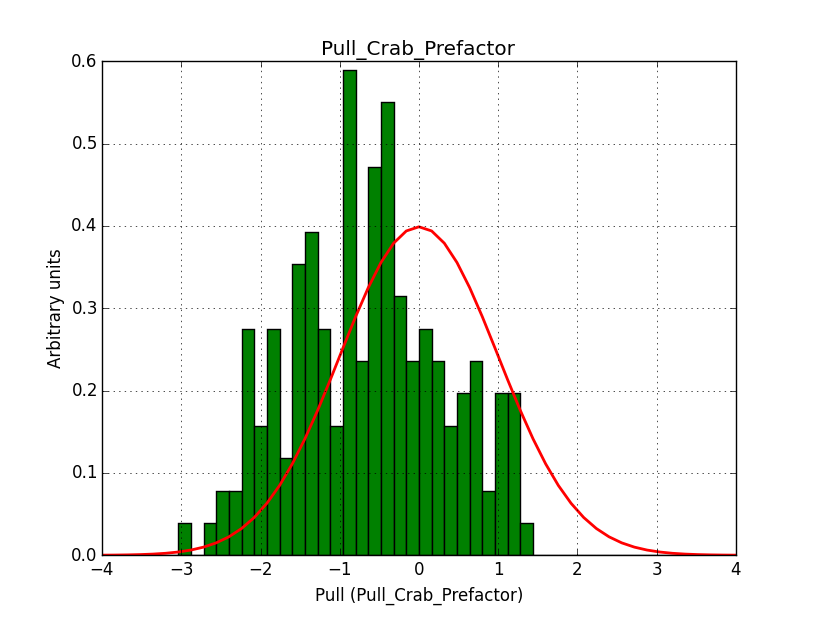
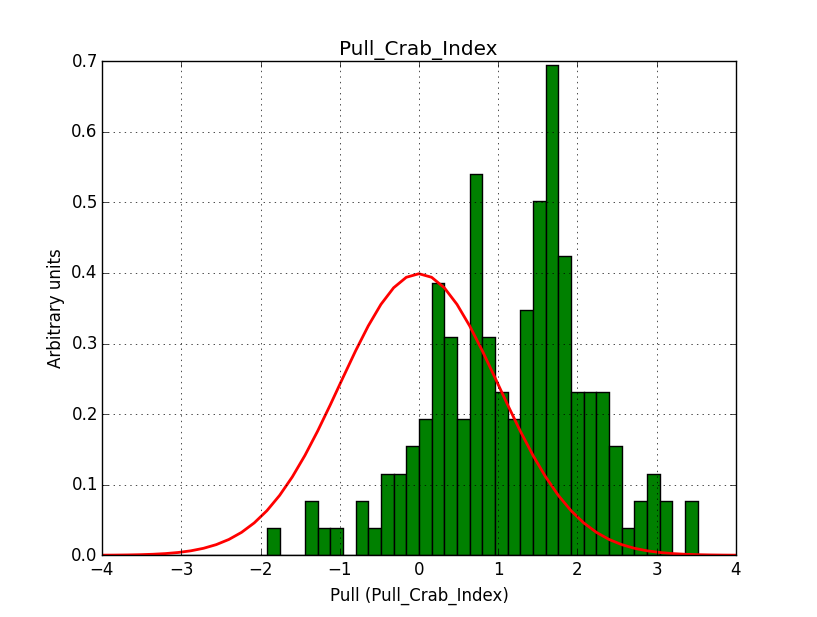
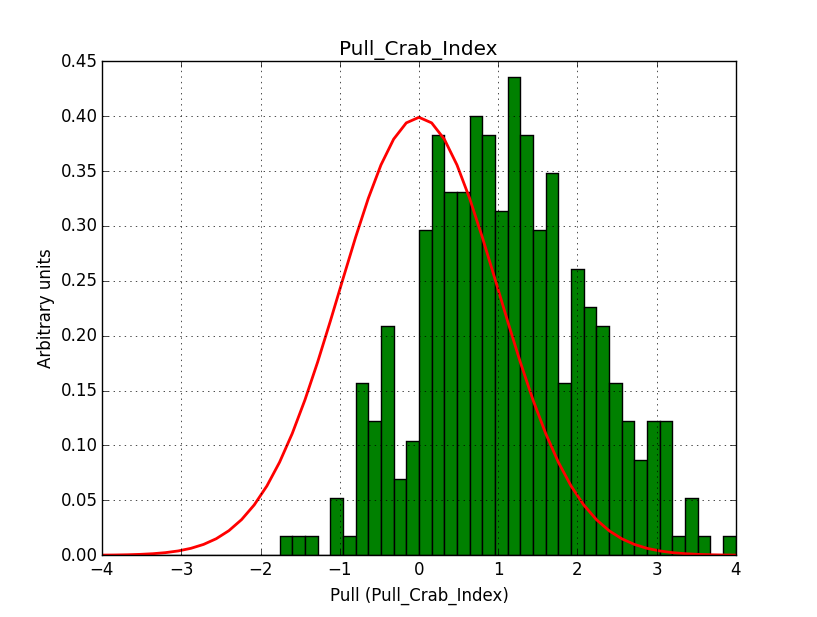
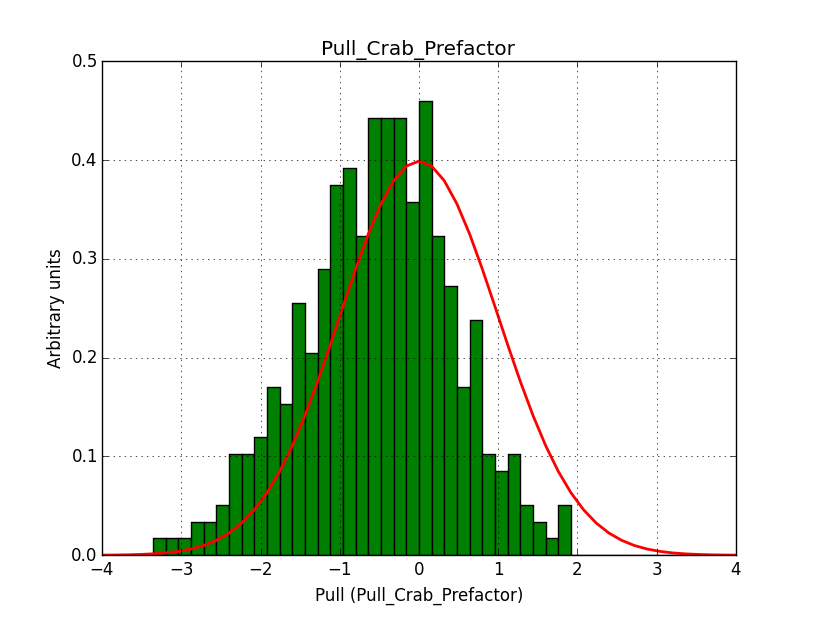
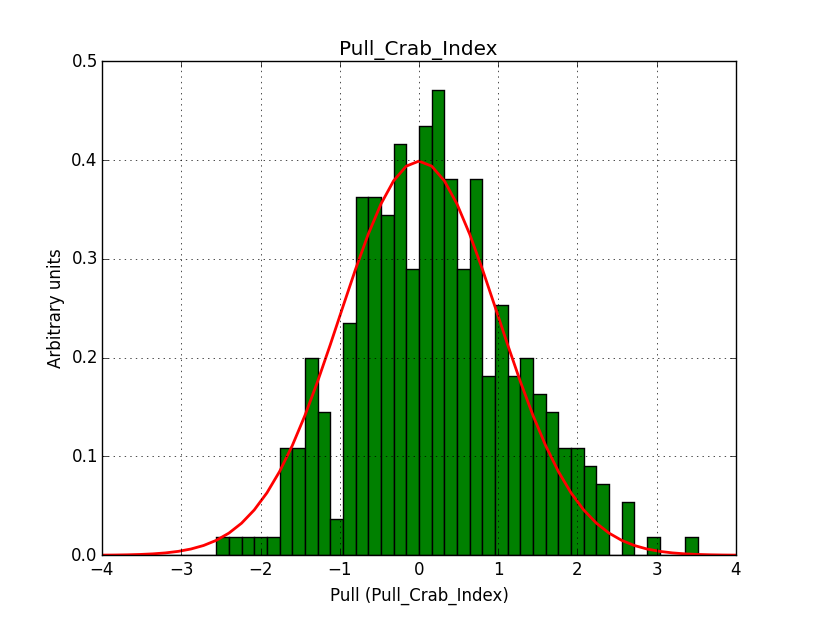
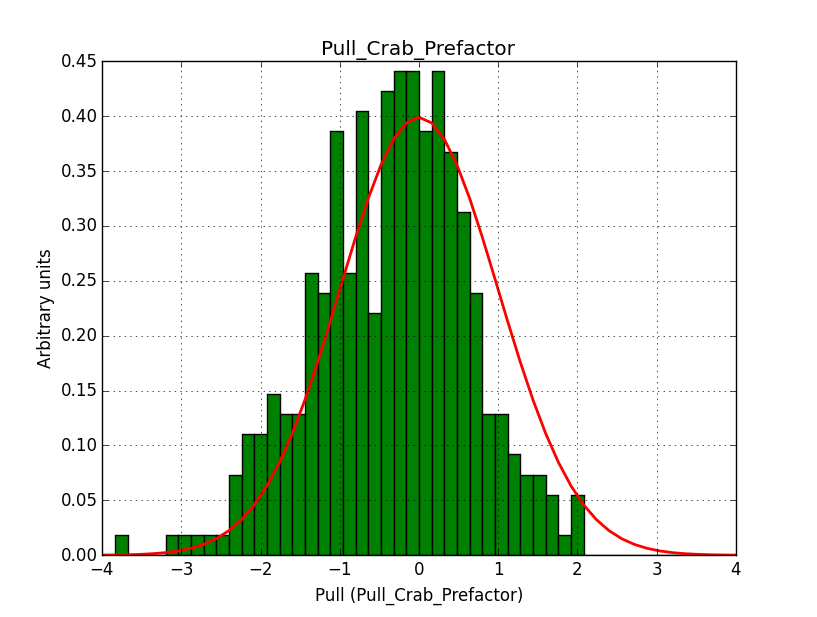
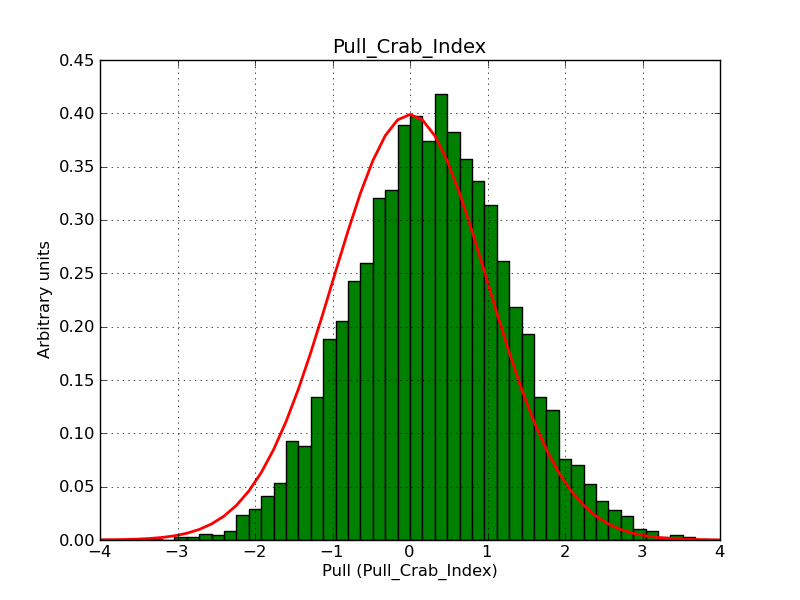
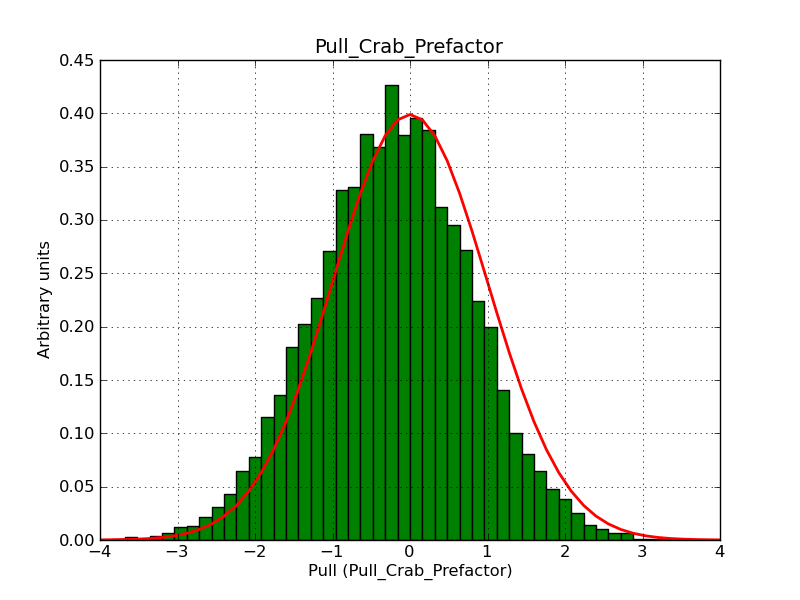
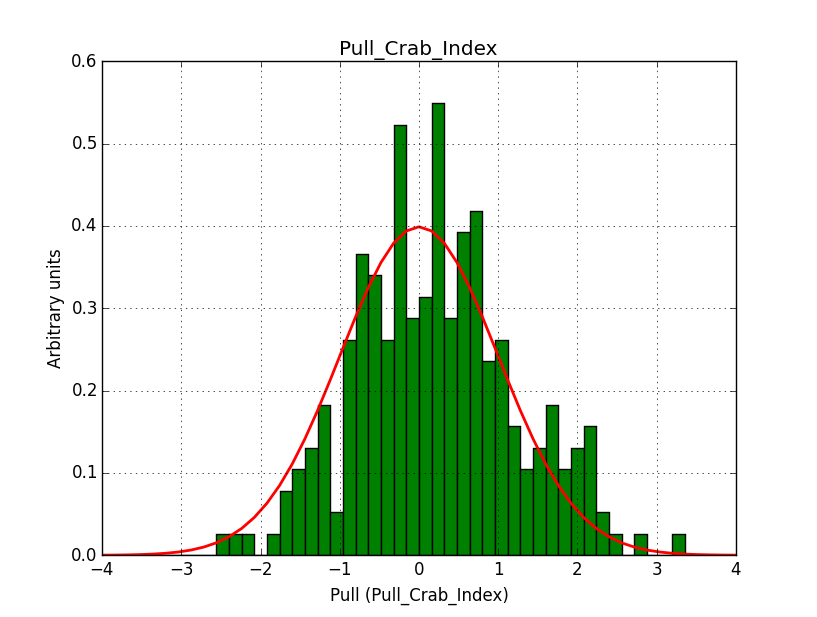
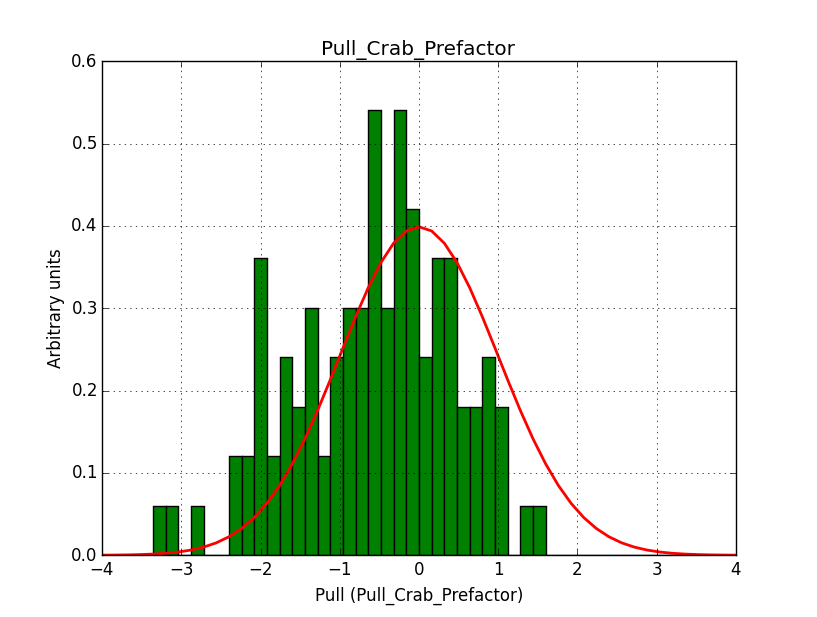
I modyfied cspull to handle stacked observations. Using it to simulate and fit three observations, containing the Crab and a CTA background as sources, results in the following pull distributions:
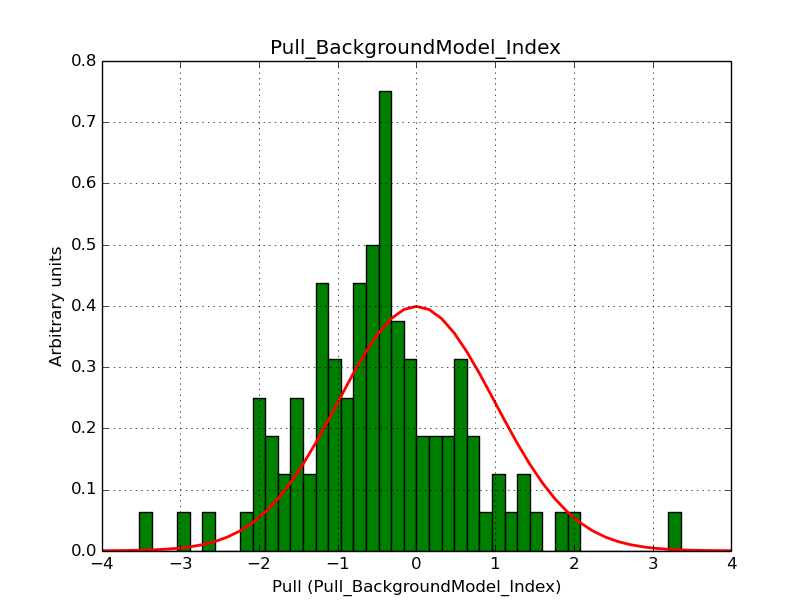
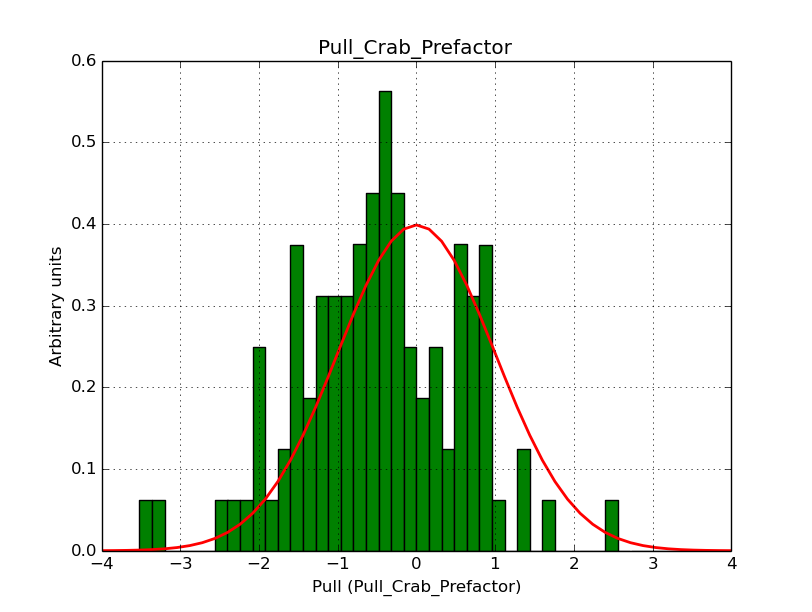
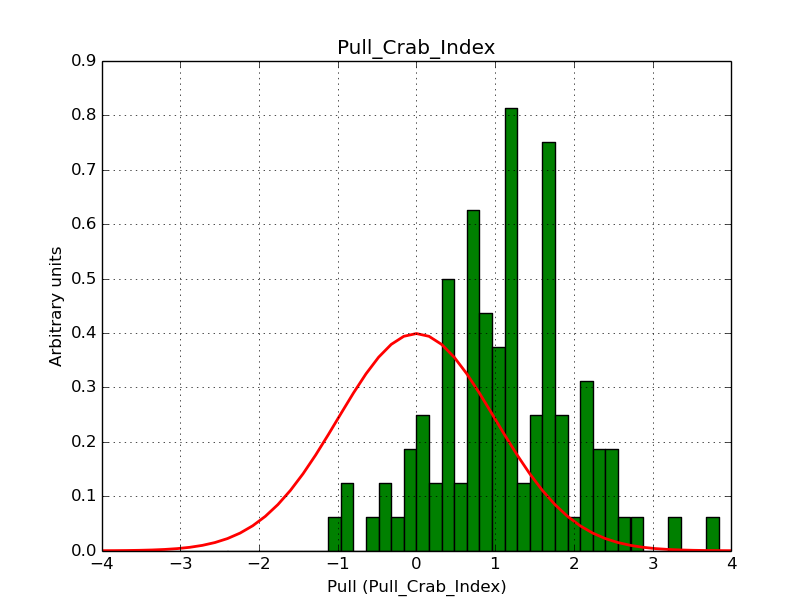
The seems to be an atribution of source photons to the background. The used observations and source models are attached. I will push the modified cspull after some small further improvements to it.
Recurrence
No recurrence.
History
#1
 Updated by Buehler Rolf over 9 years ago
Updated by Buehler Rolf over 9 years ago
#2
 Updated by Buehler Rolf over 9 years ago
Updated by Buehler Rolf over 9 years ago
- File cspull_input.png added
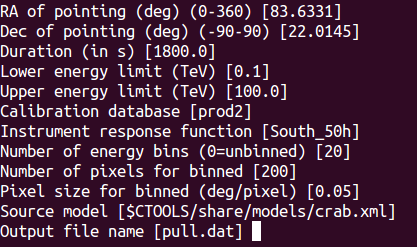
The input parameters are shown in the image shown below. Note that the expcube, psfcube, bkgcube are done with the same binning as the ctscube, which is not efficient, but avoids user errors.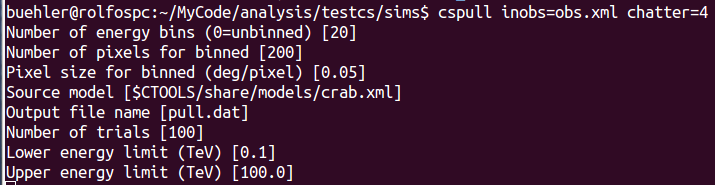
#3
 Updated by Buehler Rolf over 9 years ago
Updated by Buehler Rolf over 9 years ago
- File cspull_input.png added
#4
 Updated by Buehler Rolf over 9 years ago
Updated by Buehler Rolf over 9 years ago
- File cspull_input.png added
- File Pull_BackgroundModel_Index.png added
- File Pull_BackgroundModel_Prefactor.png added
- File Pull_Crab_Index.png added
- File Pull_Crab_Prefactor.png added
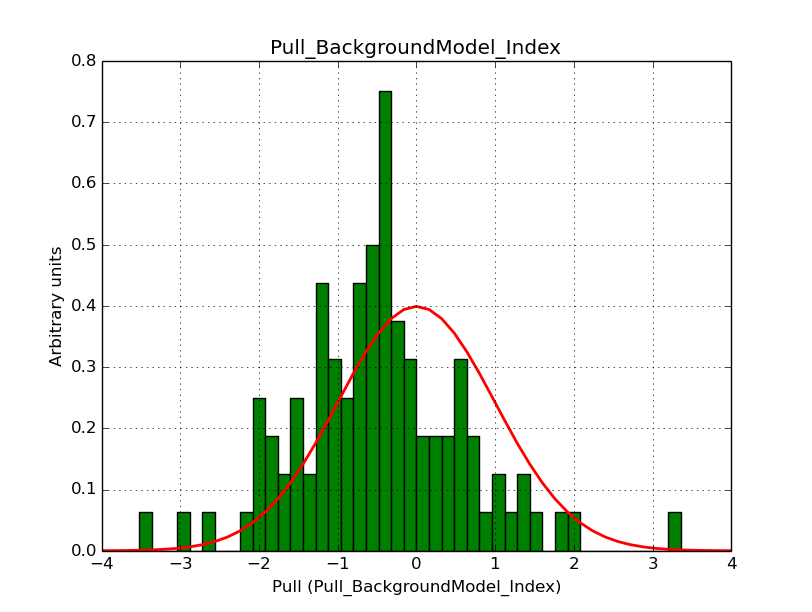
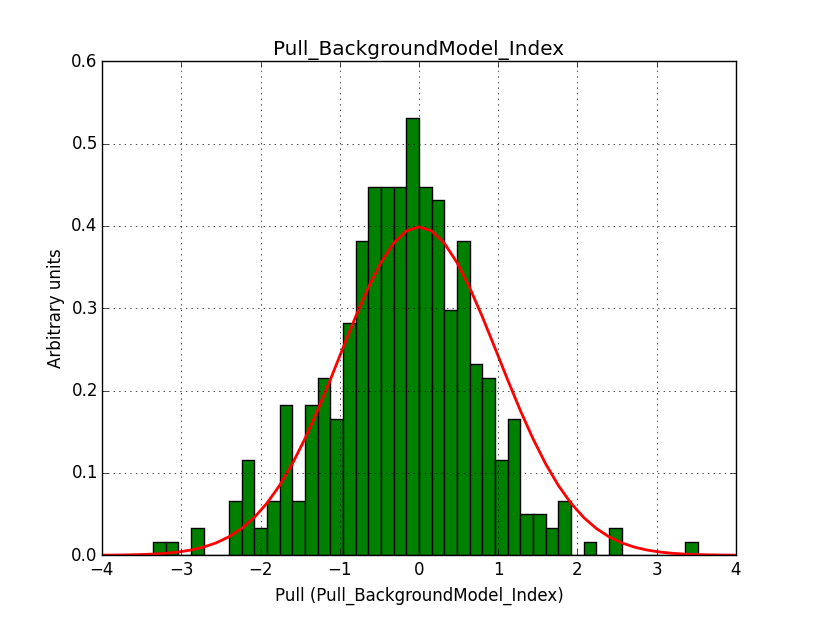
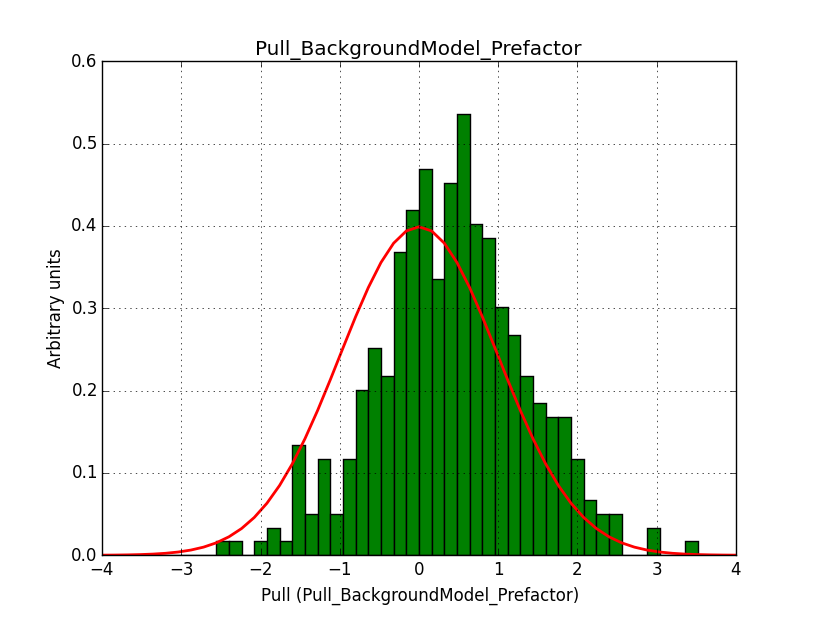
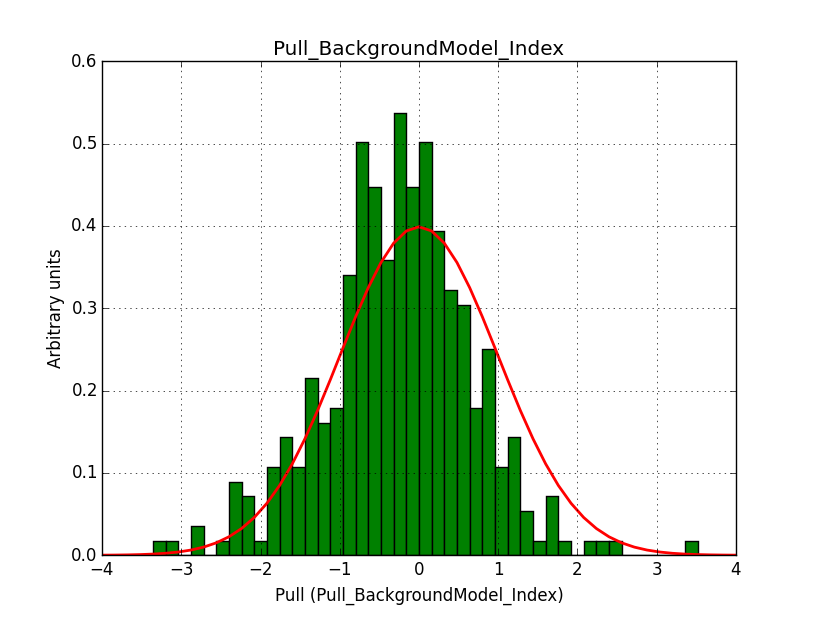
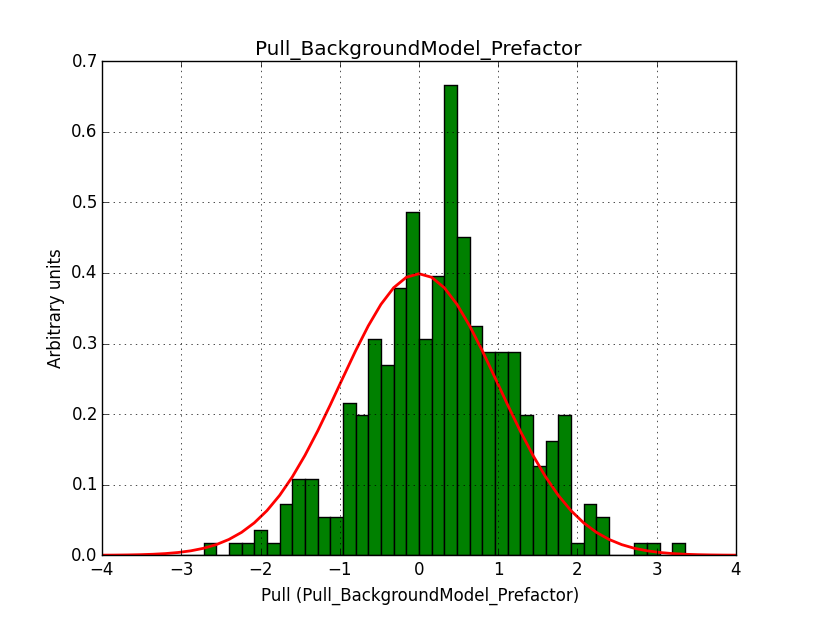
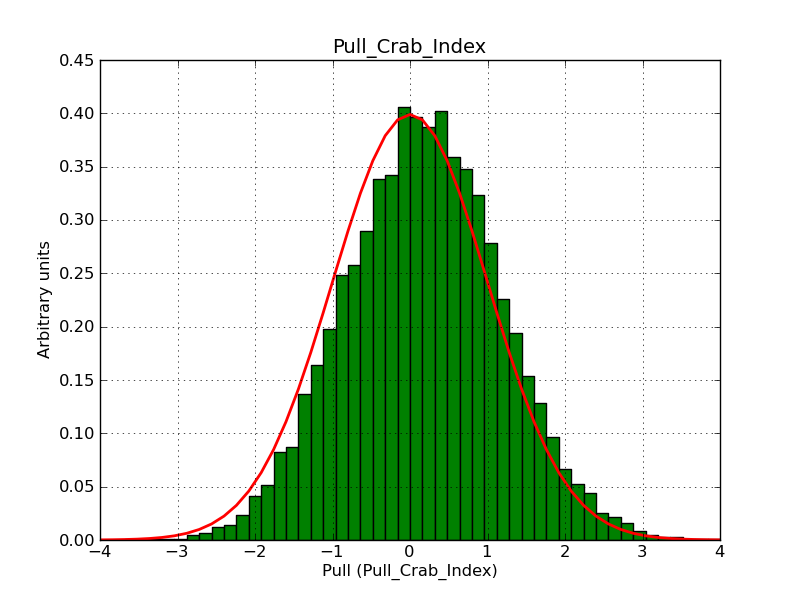
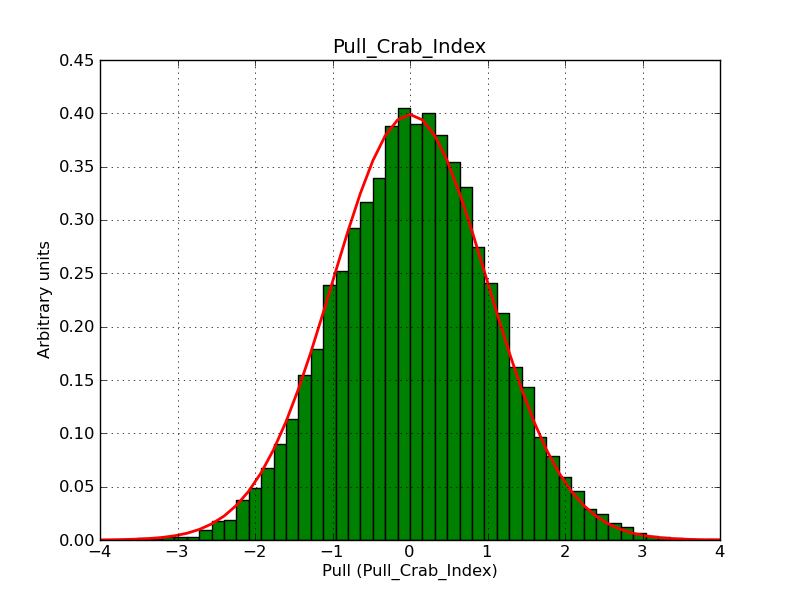
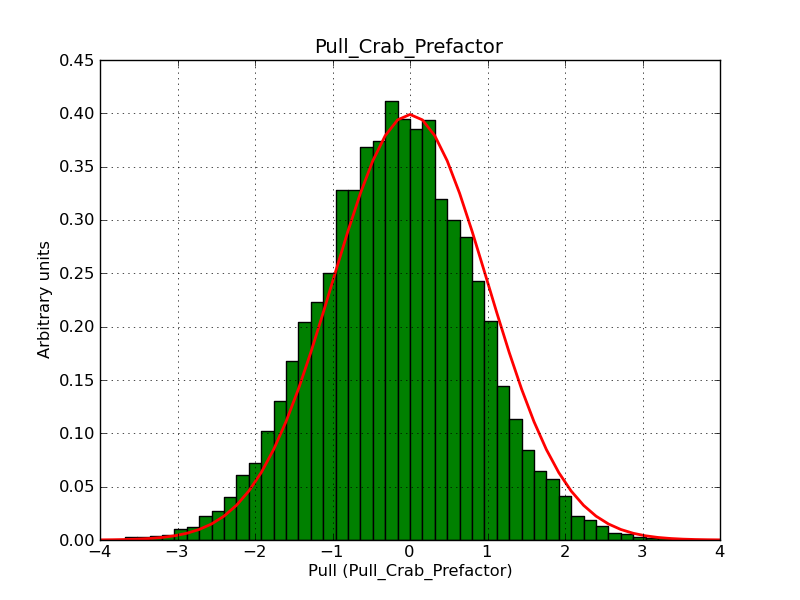
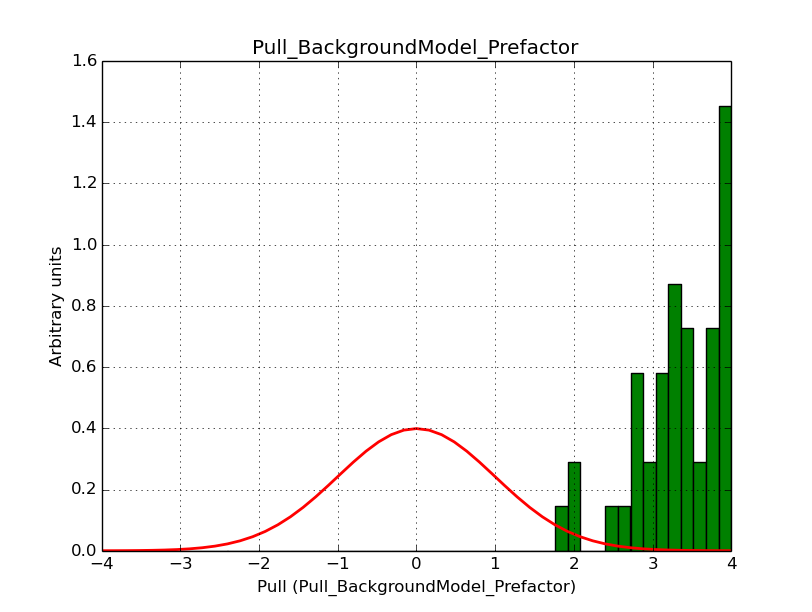
I fixed a bug in my cspull version. This changed the distributions, but they are now even more offset for the background (I updated the figures, so the plots above show the correct plots). I will push my cspull modifications into devel.
#5
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- File pull_b20_prefactor.png added
- File pull_b20_index.png added
- File pull_b40_index.png added
- File pull_b40_prefactor.png added
- File pull_b40_bkg_prefactor.png added
- File pull_b40_bkg_index.png added
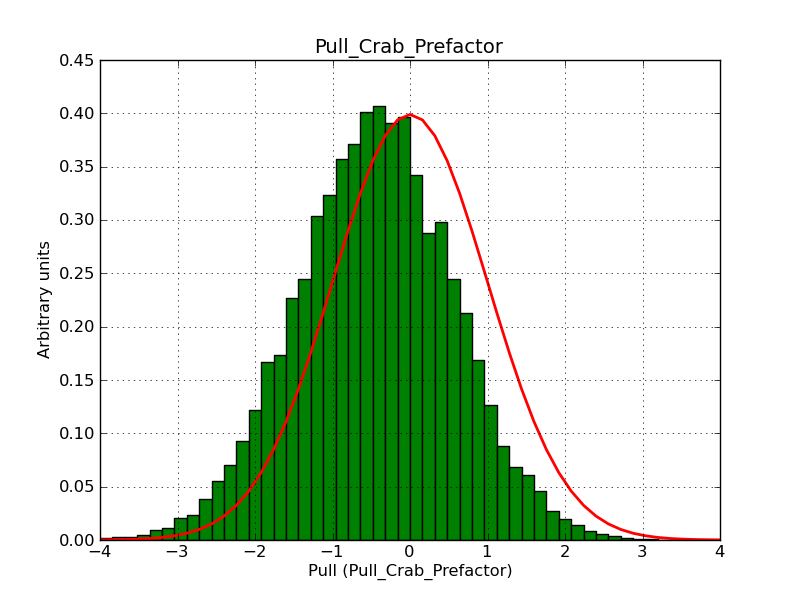
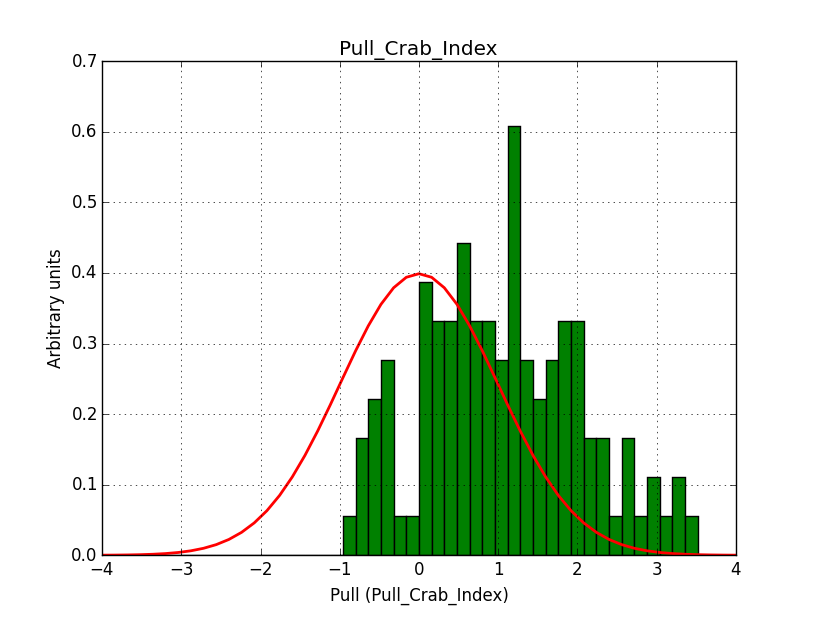
I integrated the modified cspull script into ctools. Please note that I have removed the expcube, psfcube and bkgcube parameters from cspull as they were in fact not used. You need to clean-up your pfiles folder so that you don’t have an old version with these parameters lurking around.
It turned out that the background cube model indeed had the livetime correction twice. The background cube rates are counts/livetime, but then there was an additional correction in the GCTAModelCubeBackground code. I removed that. The background pulls are now much more reasonable.
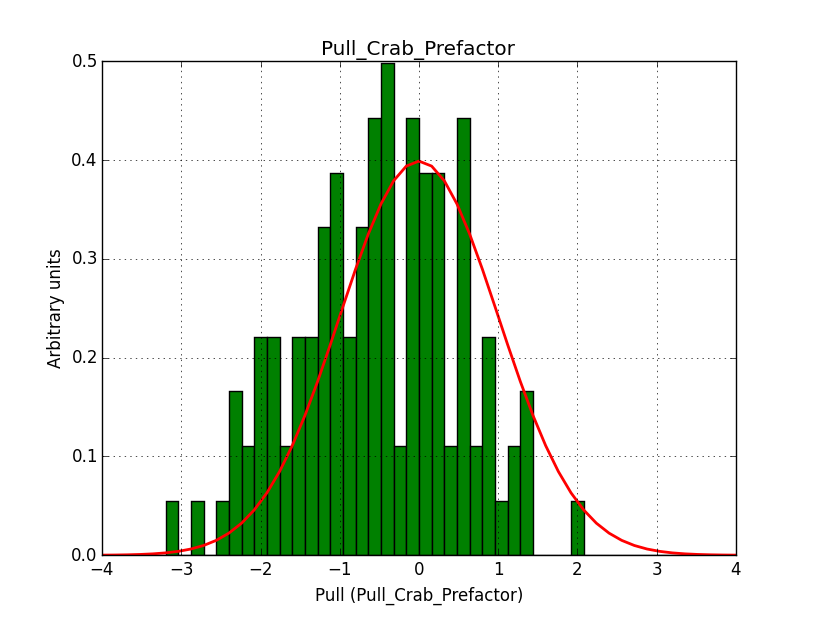 |
 |
 |
 |
I also provide for reference the background model spectral parameter pull histograms for 40 bins.
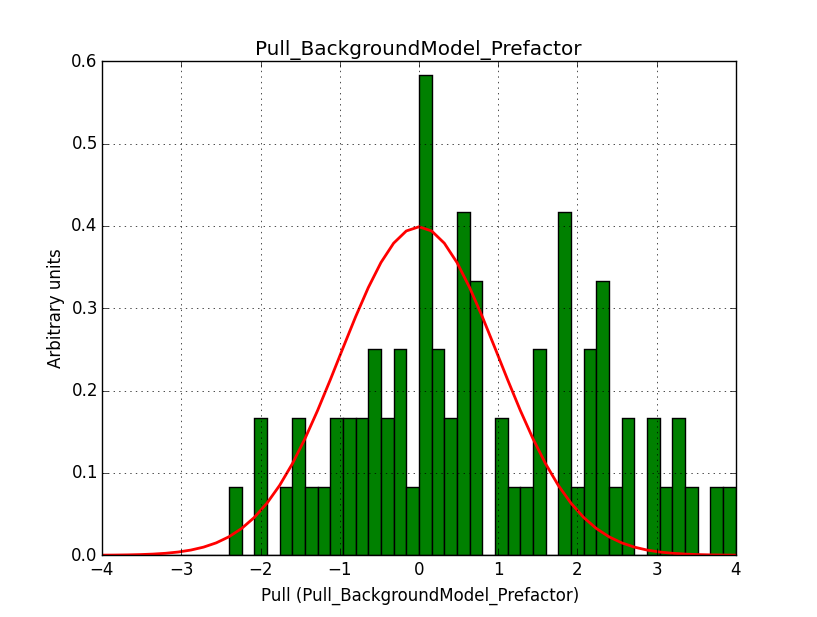 |
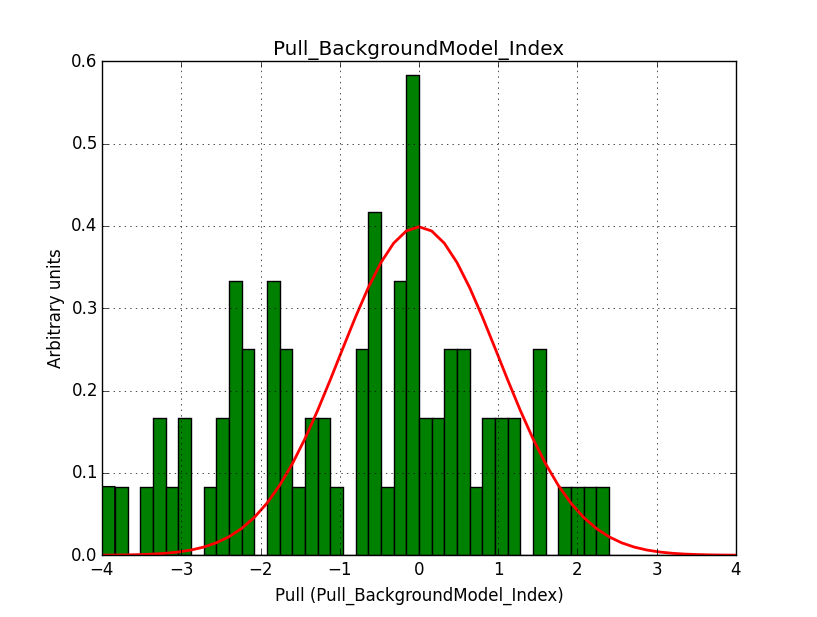 |
Note that the fitting shows always a residual in Npred. Not clear whether this is a bad Npred computation, or whether this is actually a problem in the fit (which seems to converge):
2015-07-03T14:10:40: +=================================+ 2015-07-03T14:10:40: | Maximum likelihood optimisation | 2015-07-03T14:10:40: +=================================+ 2015-07-03T14:10:41: >Iteration 0: -logL=47448.077, Lambda=1.0e-03 2015-07-03T14:10:42: >Iteration 1: -logL=47447.780, Lambda=1.0e-03, delta=0.298, max(|grad|)=7186008816.917294 [Index:7] 2015-07-03T14:10:43: >Iteration 2: -logL=47447.779, Lambda=1.0e-04, delta=0.000, max(|grad|)=7186004339.309735 [Index:7] 2015-07-03T14:10:43: 2015-07-03T14:10:43: +==================+ 2015-07-03T14:10:43: | Curvature matrix | 2015-07-03T14:10:43: +==================+ 2015-07-03T14:10:43: === GMatrixSparse === 2015-07-03T14:10:43: Number of rows ............: 10 2015-07-03T14:10:43: Number of columns .........: 10 2015-07-03T14:10:43: Number of nonzero elements : 16 2015-07-03T14:10:43: Number of allocated cells .: 528 2015-07-03T14:10:43: Memory block size .........: 512 2015-07-03T14:10:43: Sparse matrix fill ........: 0.16 2015-07-03T14:10:43: Pending element ...........: 0 2015-07-03T14:10:43: Fill stack size ...........: 0 (none) 2015-07-03T14:10:43: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T14:10:43: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T14:10:43: 0, 0, 188.146, -286.823, 0, 0, 1.12635e+06, 3.47759e+06, 0, 0 2015-07-03T14:10:43: 0, 0, -286.823, 8616.04, 0, 0, -2.7315e+07, -9.1871e+07, 0, 0 2015-07-03T14:10:43: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T14:10:43: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T14:10:43: 0, 0, 1.12635e+06, -2.7315e+07, 0, 0, 1.63675e+17, 5.60271e+17, 0, 0 2015-07-03T14:10:43: 0, 0, 3.47759e+06, -9.1871e+07, 0, 0, 5.60271e+17, 1.93156e+18, 0, 0 2015-07-03T14:10:43: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T14:10:43: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T14:10:43: 2015-07-03T14:10:43: +=========================================+ 2015-07-03T14:10:43: | Maximum likelihood optimization results | 2015-07-03T14:10:43: +=========================================+ 2015-07-03T14:10:43: === GOptimizerLM === 2015-07-03T14:10:43: Optimized function value ..: 47447.779 2015-07-03T14:10:43: Absolute precision ........: 0.005 2015-07-03T14:10:43: Acceptable value decrease .: 2 2015-07-03T14:10:43: Optimization status .......: converged 2015-07-03T14:10:43: Number of parameters ......: 10 2015-07-03T14:10:43: Number of free parameters .: 4 2015-07-03T14:10:43: Number of iterations ......: 2 2015-07-03T14:10:43: Lambda ....................: 1e-05 2015-07-03T14:10:43: Maximum log likelihood ....: -47447.779 2015-07-03T14:10:43: Observed events (Nobs) ...: 39586.000 2015-07-03T14:10:43: Predicted events (Npred) ..: 37661.415 (Nobs - Npred = 1924.59)
#6
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- File pull_b20_prefactor_binsz002.png added
- File pull_b20_index_binsz002.png added
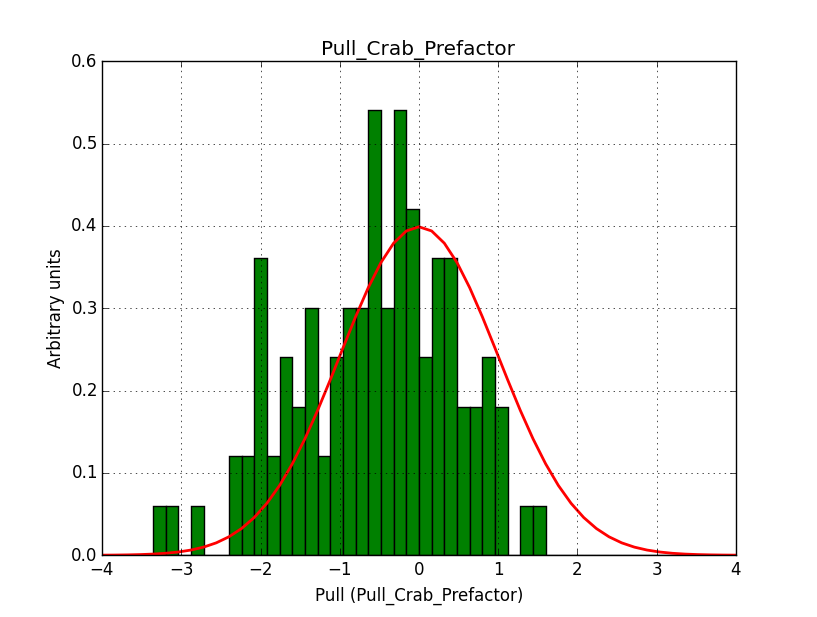
There is a dependence on the size of the counts map. The original runs were for a 100 x 100 pixel map with 0.05 deg pixels, hence covering an area of 5 x 5 deg.
I tried a smaller map with a binsize of 0.02 deg and still 100 x 100 pixels, covering 2 x 2 deg.
 |
 |
There is still an issue with the Npred value:
2015-07-03T16:02:29: +=========================================+ 2015-07-03T16:02:29: | Maximum likelihood optimization results | 2015-07-03T16:02:29: +=========================================+ 2015-07-03T16:02:29: === GOptimizerLM === 2015-07-03T16:02:29: Optimized function value ..: 16666.857 2015-07-03T16:02:29: Absolute precision ........: 0.005 2015-07-03T16:02:29: Acceptable value decrease .: 2 2015-07-03T16:02:29: Optimization status .......: converged 2015-07-03T16:02:29: Number of parameters ......: 10 2015-07-03T16:02:29: Number of free parameters .: 4 2015-07-03T16:02:29: Number of iterations ......: 15 2015-07-03T16:02:29: Lambda ....................: 0.01 2015-07-03T16:02:29: Maximum log likelihood ....: -16666.857 2015-07-03T16:02:29: Observed events (Nobs) ...: 16452.000 2015-07-03T16:02:29: Predicted events (Npred) ..: 15032.236 (Nobs - Npred = 1419.76)
#7
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- Status changed from New to In Progress
- Assigned To set to Knödlseder Jürgen
- % Done changed from 0 to 10
#8
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
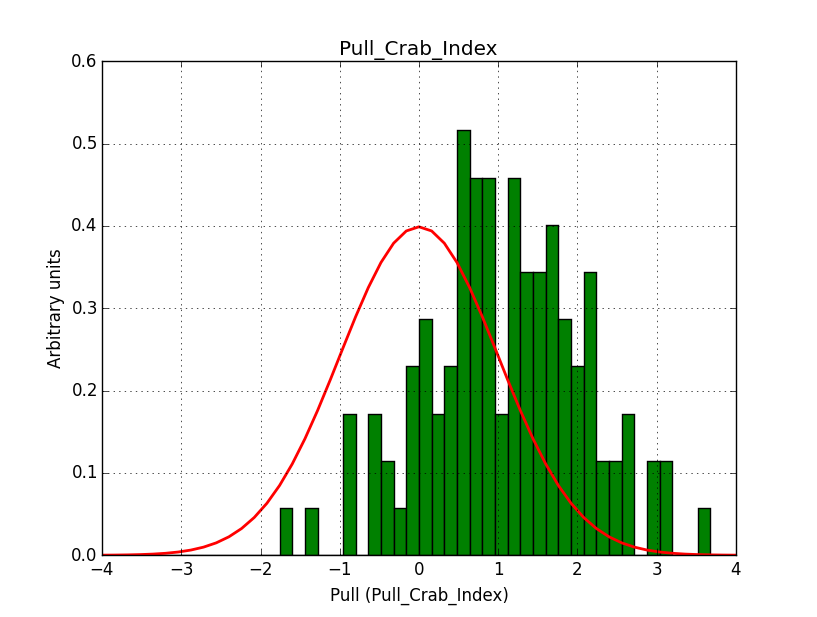
I did another test without offset between the runs, hence 3 observations with the same pointing (Crab) and an ROI of 5 deg.
I used 20 bins in energy and 100 x 100 pixels of 0.05 deg/pixel, i.e. a 5 x 5 deg counts map. Below the resulting pull distribution. There is still the bias in the index. This seems to be inherent to large maps.
 |
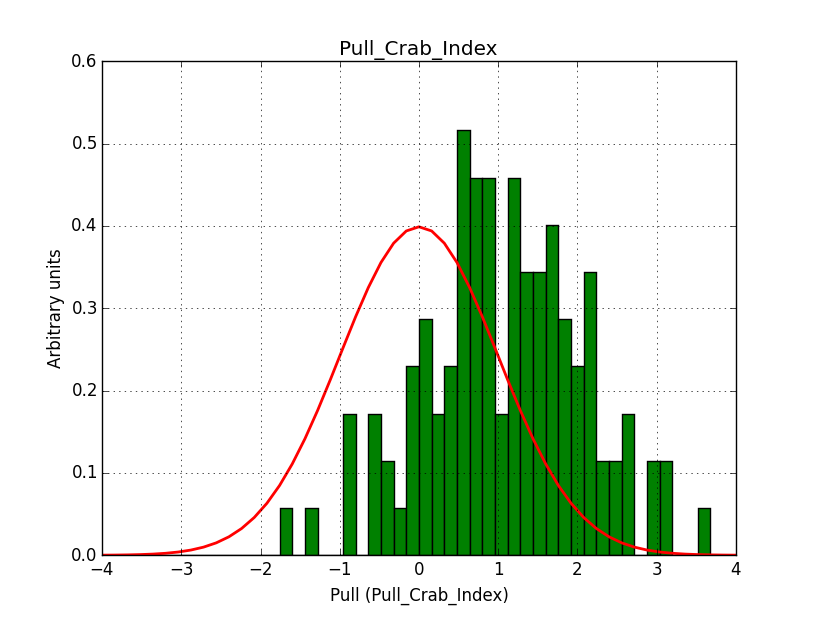 |
The Npred value is also too large:
2015-07-03T16:23:52: === GOptimizerLM === 2015-07-03T16:23:52: Optimized function value ..: 10240.417 2015-07-03T16:23:52: Absolute precision ........: 0.005 2015-07-03T16:23:52: Acceptable value decrease .: 2 2015-07-03T16:23:52: Optimization status .......: converged 2015-07-03T16:23:52: Number of parameters ......: 10 2015-07-03T16:23:52: Number of free parameters .: 4 2015-07-03T16:23:52: Number of iterations ......: 6 2015-07-03T16:23:52: Lambda ....................: 0.1 2015-07-03T16:23:52: Maximum log likelihood ....: -10240.417 2015-07-03T16:23:52: Observed events (Nobs) ...: 45473.000 2015-07-03T16:23:52: Predicted events (Npred) ..: 43741.983 (Nobs - Npred = 1731.02)
#9
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- File pull_b20_id_index.png added
- File pull_b20_id_prefactor.png added
#10
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- File pull_b20_single_prefactor.png added
- File pull_b20_single_index.png added
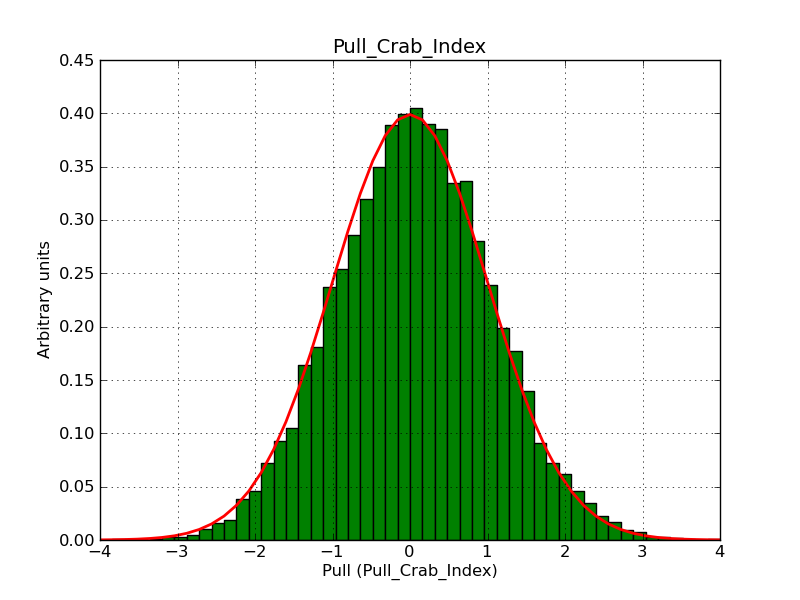
For comparison, I did the same simulation, but now using a classical binned analysis. The difference is now that the response information is taken from the original IRFs, and not the stacked versions. The results show the same shifts, but the Npred number is now correct. Note that the fit gradients are also much smaller.
I’m wondering whether there are in fact two different problems to understand.
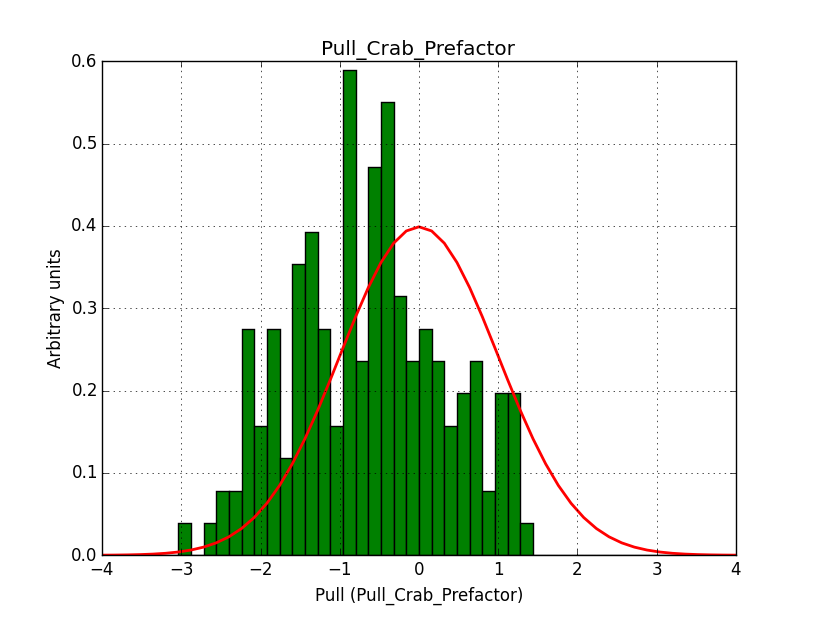 |
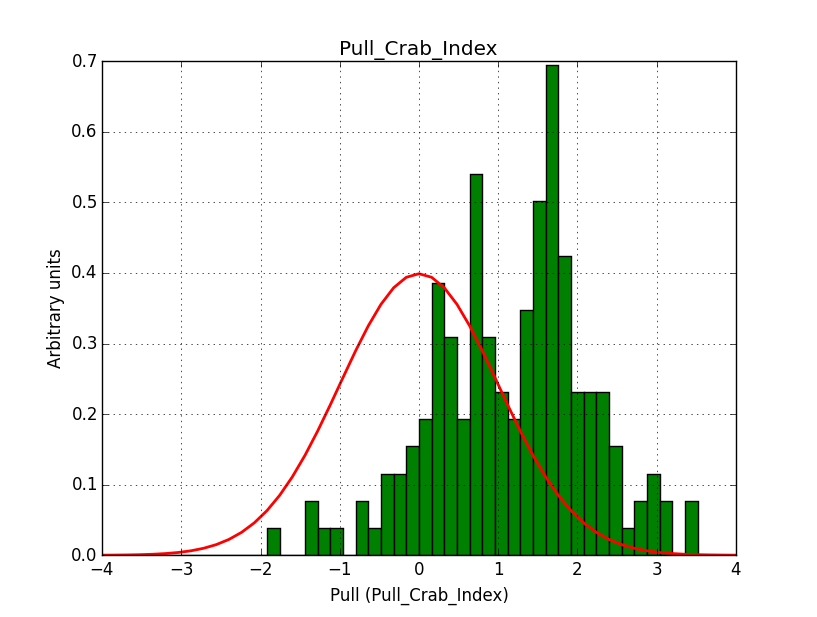 |
2015-07-03T16:46:08: +=================================+ 2015-07-03T16:46:08: | Maximum likelihood optimisation | 2015-07-03T16:46:08: +=================================+ 2015-07-03T16:46:09: >Iteration 0: -logL=14855.667, Lambda=1.0e-03 2015-07-03T16:46:09: >Iteration 1: -logL=14851.929, Lambda=1.0e-03, delta=3.738, max(|grad|)=4.217249 [Index:7] 2015-07-03T16:46:10: >Iteration 2: -logL=14851.928, Lambda=1.0e-04, delta=0.001, max(|grad|)=0.027799 [Index:3] 2015-07-03T16:46:10: 2015-07-03T16:46:10: +==================+ 2015-07-03T16:46:10: | Curvature matrix | 2015-07-03T16:46:10: +==================+ 2015-07-03T16:46:10: === GMatrixSparse === 2015-07-03T16:46:10: Number of rows ............: 10 2015-07-03T16:46:10: Number of columns .........: 10 2015-07-03T16:46:10: Number of nonzero elements : 16 2015-07-03T16:46:10: Number of allocated cells .: 528 2015-07-03T16:46:10: Memory block size .........: 512 2015-07-03T16:46:10: Sparse matrix fill ........: 0.16 2015-07-03T16:46:10: Pending element ...........: 0 2015-07-03T16:46:10: Fill stack size ...........: 0 (none) 2015-07-03T16:46:10: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T16:46:10: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T16:46:10: 0, 0, 259.335, -320.444, 0, 0, 82.9803, -147.697, 0, 0 2015-07-03T16:46:10: 0, 0, -320.444, 10997.4, 0, 0, 264.36, -593.486, 0, 0 2015-07-03T16:46:10: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T16:46:10: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T16:46:10: 0, 0, 82.9803, 264.36, 0, 0, 35867.2, -58400.5, 0, 0 2015-07-03T16:46:10: 0, 0, -147.697, -593.486, 0, 0, -58400.5, 110337, 0, 0 2015-07-03T16:46:10: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T16:46:10: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T16:46:11: 2015-07-03T16:46:11: +=========================================+ 2015-07-03T16:46:11: | Maximum likelihood optimization results | 2015-07-03T16:46:11: +=========================================+ 2015-07-03T16:46:11: === GOptimizerLM === 2015-07-03T16:46:11: Optimized function value ..: 14851.928 2015-07-03T16:46:11: Absolute precision ........: 0.005 2015-07-03T16:46:11: Acceptable value decrease .: 2 2015-07-03T16:46:11: Optimization status .......: converged 2015-07-03T16:46:11: Number of parameters ......: 10 2015-07-03T16:46:11: Number of free parameters .: 4 2015-07-03T16:46:11: Number of iterations ......: 2 2015-07-03T16:46:11: Lambda ....................: 1e-05 2015-07-03T16:46:11: Maximum log likelihood ....: -14851.928 2015-07-03T16:46:11: Observed events (Nobs) ...: 45144.000 2015-07-03T16:46:11: Predicted events (Npred) ..: 45143.998 (Nobs - Npred = 0.0015177)
#11
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- File residual.fits.gz added
I checked the difference between a model map computed from the IRFs and a model map computed from the cube response, for the same input parameters and observation definition (1800 seconds, Crab as a single source).
I used the following script to compute the residual map:
import gammalib
map_irf = gammalib.GSkyMap("modcube_irf.fits")
map_stacked = gammalib.GSkyMap("modcube_stacked.fits")
residual = (map_irf - map_stacked) / map_irf
residual.save("residual.fits", True)
There is a ring-like residual at the position of the Crab, but the amplitude of this residual in individual pixels does not exceed 1-2%. Attached the generated FITS file (residual.fits.gz). The response computation is thus consistent.
#12
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- % Done changed from 10 to 50
I finally found the bug in the Npred computation: when removing the deadtime correction I also removed some renormalization of the gradients that was needed to get their right amplitude. The gradients of the background model were thus wrong. This is now corrected.
For reference, here the fit when using the IRF computation for the binned analysis:
2015-07-03T21:32:26: +=================================+ 2015-07-03T21:32:26: | Maximum likelihood optimisation | 2015-07-03T21:32:26: +=================================+ 2015-07-03T21:32:28: >Iteration 0: -logL=39901.606, Lambda=1.0e-03 2015-07-03T21:32:29: >Iteration 1: -logL=39899.679, Lambda=1.0e-03, delta=1.928, max(|grad|)=6.256420 [Index:7] 2015-07-03T21:32:31: >Iteration 2: -logL=39899.678, Lambda=1.0e-04, delta=0.001, max(|grad|)=-0.059539 [Index:3] 2015-07-03T21:32:31: 2015-07-03T21:32:31: +==================+ 2015-07-03T21:32:31: | Curvature matrix | 2015-07-03T21:32:31: +==================+ 2015-07-03T21:32:31: === GMatrixSparse === 2015-07-03T21:32:31: Number of rows ............: 10 2015-07-03T21:32:31: Number of columns .........: 10 2015-07-03T21:32:31: Number of nonzero elements : 16 2015-07-03T21:32:31: Number of allocated cells .: 528 2015-07-03T21:32:31: Memory block size .........: 512 2015-07-03T21:32:31: Sparse matrix fill ........: 0.16 2015-07-03T21:32:31: Pending element ...........: 0 2015-07-03T21:32:31: Fill stack size ...........: 0 (none) 2015-07-03T21:32:31: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:32:31: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:32:31: 0, 0, 83.7417, -95.0523, 0, 0, 25.4407, -46.9068, 0, 0 2015-07-03T21:32:31: 0, 0, -95.0523, 3812.11, 0, 0, 89.0835, -199.93, 0, 0 2015-07-03T21:32:31: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:32:31: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:32:31: 0, 0, 25.4407, 89.0835, 0, 0, 9879.25, -16499.1, 0, 0 2015-07-03T21:32:31: 0, 0, -46.9068, -199.93, 0, 0, -16499.1, 31620.7, 0, 0 2015-07-03T21:32:31: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:32:31: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:32:32: 2015-07-03T21:32:32: +=========================================+ 2015-07-03T21:32:32: | Maximum likelihood optimization results | 2015-07-03T21:32:32: +=========================================+ 2015-07-03T21:32:32: === GOptimizerLM === 2015-07-03T21:32:32: Optimized function value ..: 39899.678 2015-07-03T21:32:32: Absolute precision ........: 0.005 2015-07-03T21:32:32: Acceptable value decrease .: 2 2015-07-03T21:32:32: Optimization status .......: converged 2015-07-03T21:32:32: Number of parameters ......: 10 2015-07-03T21:32:32: Number of free parameters .: 4 2015-07-03T21:32:32: Number of iterations ......: 2 2015-07-03T21:32:32: Lambda ....................: 1e-05 2015-07-03T21:32:32: Maximum log likelihood ....: -39899.678 2015-07-03T21:32:32: Observed events (Nobs) ...: 13359.000 2015-07-03T21:32:32: Predicted events (Npred) ..: 13358.998 (Nobs - Npred = 0.00151521) 2015-07-03T21:32:32: === GModels === 2015-07-03T21:32:32: Number of models ..........: 2 2015-07-03T21:32:32: Number of parameters ......: 10 2015-07-03T21:32:32: === GModelSky === 2015-07-03T21:32:32: Name ......................: Crab 2015-07-03T21:32:32: Instruments ...............: all 2015-07-03T21:32:32: Instrument scale factors ..: unity 2015-07-03T21:32:32: Observation identifiers ...: all 2015-07-03T21:32:32: Model type ................: PointSource 2015-07-03T21:32:32: Model components ..........: "SkyDirFunction" * "PowerLaw" * "Constant" 2015-07-03T21:32:32: Number of parameters ......: 6 2015-07-03T21:32:32: Number of spatial par's ...: 2 2015-07-03T21:32:32: RA .......................: 83.6331 [-360,360] deg (fixed,scale=1) 2015-07-03T21:32:32: DEC ......................: 22.0145 [-90,90] deg (fixed,scale=1) 2015-07-03T21:32:32: Number of spectral par's ..: 3 2015-07-03T21:32:32: Prefactor ................: 5.78359e-16 +/- 1.10915e-17 [1e-23,1e-13] ph/cm2/s/MeV (free,scale=1e-16,gradient) 2015-07-03T21:32:32: Index ....................: -2.49196 +/- 0.0164354 [-0,-5] (free,scale=-1,gradient) 2015-07-03T21:32:32: PivotEnergy ..............: 300000 [10000,1e+09] MeV (fixed,scale=1e+06,gradient) 2015-07-03T21:32:32: Number of temporal par's ..: 1 2015-07-03T21:32:32: Normalization ............: 1 (relative value) (fixed,scale=1,gradient) 2015-07-03T21:32:32: === GCTAModelIrfBackground === 2015-07-03T21:32:32: Name ......................: CTABackgroundModel 2015-07-03T21:32:32: Instruments ...............: CTA 2015-07-03T21:32:32: Instrument scale factors ..: unity 2015-07-03T21:32:32: Observation identifiers ...: all 2015-07-03T21:32:32: Model type ................: "PowerLaw" * "Constant" 2015-07-03T21:32:32: Number of parameters ......: 4 2015-07-03T21:32:32: Number of spectral par's ..: 3 2015-07-03T21:32:32: Prefactor ................: 1.01899 +/- 0.0280579 [0.001,1000] ph/cm2/s/MeV (free,scale=1,gradient) 2015-07-03T21:32:32: Index ....................: 0.0189749 +/- 0.0156848 [-5,5] (free,scale=1,gradient) 2015-07-03T21:32:32: PivotEnergy ..............: 1e+06 [10000,1e+09] MeV (fixed,scale=1e+06,gradient) 2015-07-03T21:32:32: Number of temporal par's ..: 1 2015-07-03T21:32:32: Normalization ............: 1 (relative value) (fixed,scale=1,gradient)
And here the same for the stacked analysis (now is basically identical):
2015-07-03T21:48:48: +=================================+ 2015-07-03T21:48:48: | Maximum likelihood optimisation | 2015-07-03T21:48:48: +=================================+ 2015-07-03T21:48:49: >Iteration 0: -logL=39901.545, Lambda=1.0e-03 2015-07-03T21:48:50: >Iteration 1: -logL=39899.629, Lambda=1.0e-03, delta=1.916, max(|grad|)=6.221122 [Index:7] 2015-07-03T21:48:51: >Iteration 2: -logL=39899.628, Lambda=1.0e-04, delta=0.001, max(|grad|)=-0.060144 [Index:3] 2015-07-03T21:48:51: 2015-07-03T21:48:51: +==================+ 2015-07-03T21:48:51: | Curvature matrix | 2015-07-03T21:48:51: +==================+ 2015-07-03T21:48:51: === GMatrixSparse === 2015-07-03T21:48:51: Number of rows ............: 10 2015-07-03T21:48:51: Number of columns .........: 10 2015-07-03T21:48:51: Number of nonzero elements : 16 2015-07-03T21:48:51: Number of allocated cells .: 528 2015-07-03T21:48:51: Memory block size .........: 512 2015-07-03T21:48:51: Sparse matrix fill ........: 0.16 2015-07-03T21:48:51: Pending element ...........: 0 2015-07-03T21:48:51: Fill stack size ...........: 0 (none) 2015-07-03T21:48:51: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:48:51: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:48:51: 0, 0, 83.7761, -95.0742, 0, 0, 25.3675, -46.7484, 0, 0 2015-07-03T21:48:51: 0, 0, -95.0742, 3812.14, 0, 0, 88.6885, -199.063, 0, 0 2015-07-03T21:48:51: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:48:51: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:48:51: 0, 0, 25.3675, 88.6885, 0, 0, 9881.05, -16501.8, 0, 0 2015-07-03T21:48:51: 0, 0, -46.7484, -199.063, 0, 0, -16501.8, 31624.7, 0, 0 2015-07-03T21:48:51: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:48:51: 0, 0, 0, 0, 0, 0, 0, 0, 0, 0 2015-07-03T21:48:52: 2015-07-03T21:48:52: +=========================================+ 2015-07-03T21:48:52: | Maximum likelihood optimization results | 2015-07-03T21:48:52: +=========================================+ 2015-07-03T21:48:52: === GOptimizerLM === 2015-07-03T21:48:52: Optimized function value ..: 39899.628 2015-07-03T21:48:52: Absolute precision ........: 0.005 2015-07-03T21:48:52: Acceptable value decrease .: 2 2015-07-03T21:48:52: Optimization status .......: converged 2015-07-03T21:48:52: Number of parameters ......: 10 2015-07-03T21:48:52: Number of free parameters .: 4 2015-07-03T21:48:52: Number of iterations ......: 2 2015-07-03T21:48:52: Lambda ....................: 1e-05 2015-07-03T21:48:52: Maximum log likelihood ....: -39899.628 2015-07-03T21:48:52: Observed events (Nobs) ...: 13359.000 2015-07-03T21:48:52: Predicted events (Npred) ..: 13358.998 (Nobs - Npred = 0.00150876) 2015-07-03T21:48:52: === GModels === 2015-07-03T21:48:52: Number of models ..........: 2 2015-07-03T21:48:52: Number of parameters ......: 10 2015-07-03T21:48:52: === GModelSky === 2015-07-03T21:48:52: Name ......................: Crab 2015-07-03T21:48:52: Instruments ...............: all 2015-07-03T21:48:52: Instrument scale factors ..: unity 2015-07-03T21:48:52: Observation identifiers ...: all 2015-07-03T21:48:52: Model type ................: PointSource 2015-07-03T21:48:52: Model components ..........: "SkyDirFunction" * "PowerLaw" * "Constant" 2015-07-03T21:48:52: Number of parameters ......: 6 2015-07-03T21:48:52: Number of spatial par's ...: 2 2015-07-03T21:48:52: RA .......................: 83.6331 [-360,360] deg (fixed,scale=1) 2015-07-03T21:48:52: DEC ......................: 22.0145 [-90,90] deg (fixed,scale=1) 2015-07-03T21:48:52: Number of spectral par's ..: 3 2015-07-03T21:48:52: Prefactor ................: 5.78242e-16 +/- 1.10892e-17 [1e-23,1e-13] ph/cm2/s/MeV (free,scale=1e-16,gradient) 2015-07-03T21:48:52: Index ....................: -2.49208 +/- 0.0164353 [-0,-5] (free,scale=-1,gradient) 2015-07-03T21:48:52: PivotEnergy ..............: 300000 [10000,1e+09] MeV (fixed,scale=1e+06,gradient) 2015-07-03T21:48:52: Number of temporal par's ..: 1 2015-07-03T21:48:52: Normalization ............: 1 (relative value) (fixed,scale=1,gradient) 2015-07-03T21:48:52: === GCTAModelCubeBackground === 2015-07-03T21:48:52: Name ......................: BackgroundModel 2015-07-03T21:48:52: Instruments ...............: CTA, HESS, MAGIC, VERITAS 2015-07-03T21:48:52: Instrument scale factors ..: unity 2015-07-03T21:48:52: Observation identifiers ...: all 2015-07-03T21:48:52: Model type ................: "PowerLaw" * "Constant" 2015-07-03T21:48:52: Number of parameters ......: 4 2015-07-03T21:48:52: Number of spectral par's ..: 3 2015-07-03T21:48:52: Prefactor ................: 1.01894 +/- 0.0280565 [0,infty[ ph/cm2/s/MeV (free,scale=1,gradient) 2015-07-03T21:48:52: Index ....................: 0.0189205 +/- 0.0156844 [-10,10] (free,scale=1,gradient) 2015-07-03T21:48:52: PivotEnergy ..............: 1e+06 MeV (fixed,scale=1e+06,gradient) 2015-07-03T21:48:52: Number of temporal par's ..: 1 2015-07-03T21:48:52: Normalization ............: 1 (relative value) (fixed,scale=1,gradient)
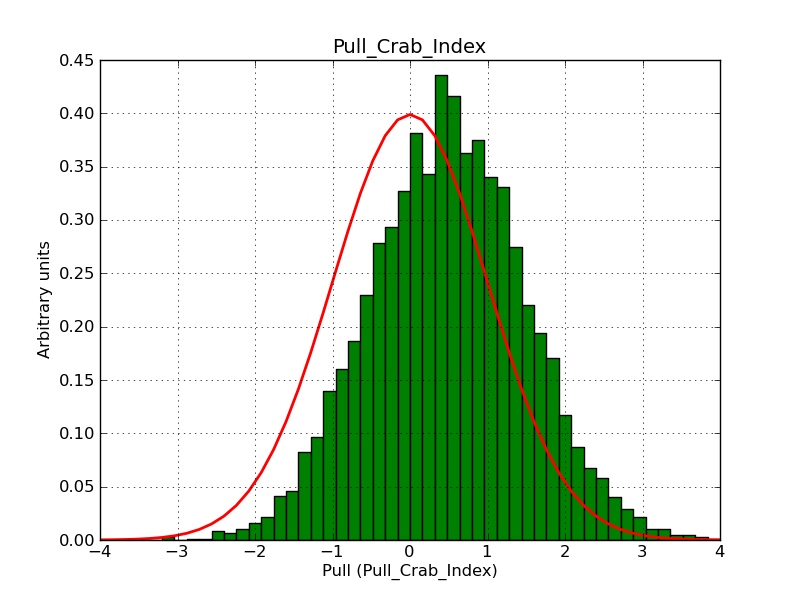
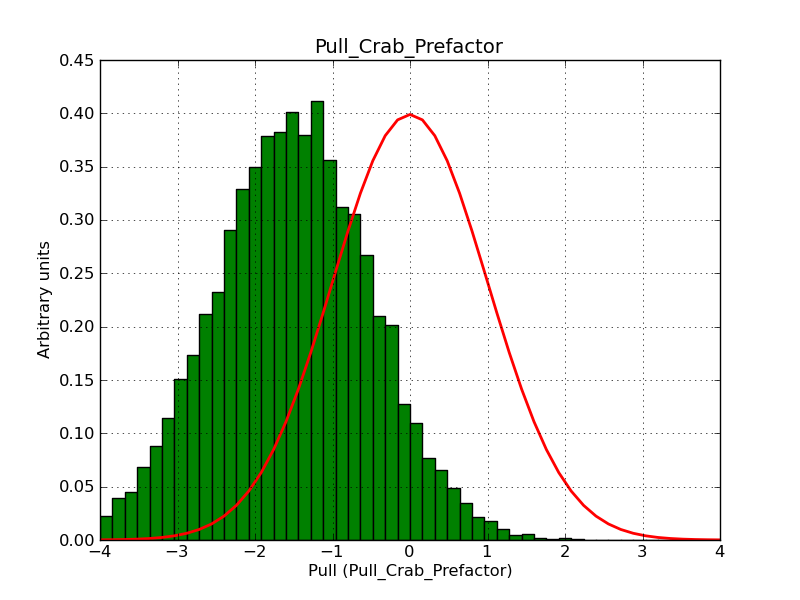
What remains to be understood is why the pull distribution of the classical binned and the stacked analyses have a bias when using a large field.
#13
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- % Done changed from 50 to 90
I understand the problem!
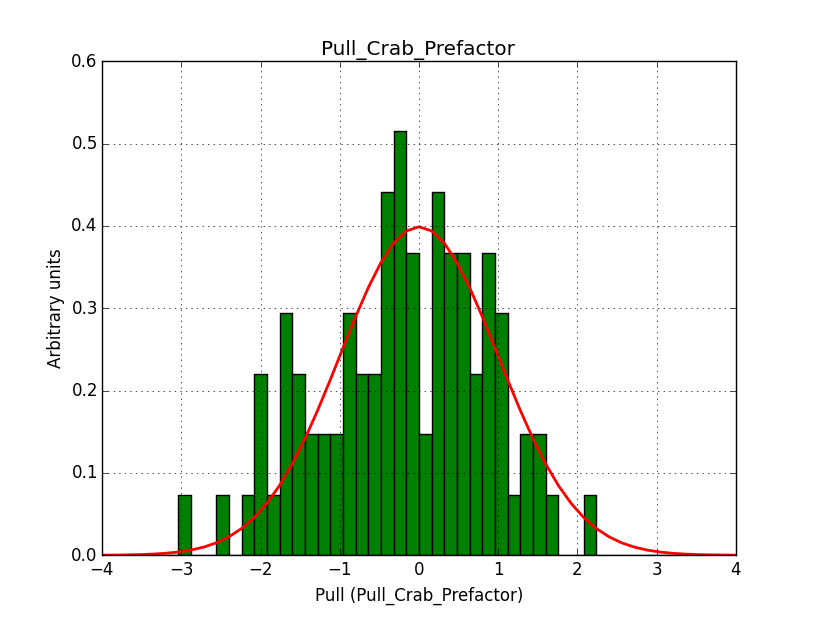
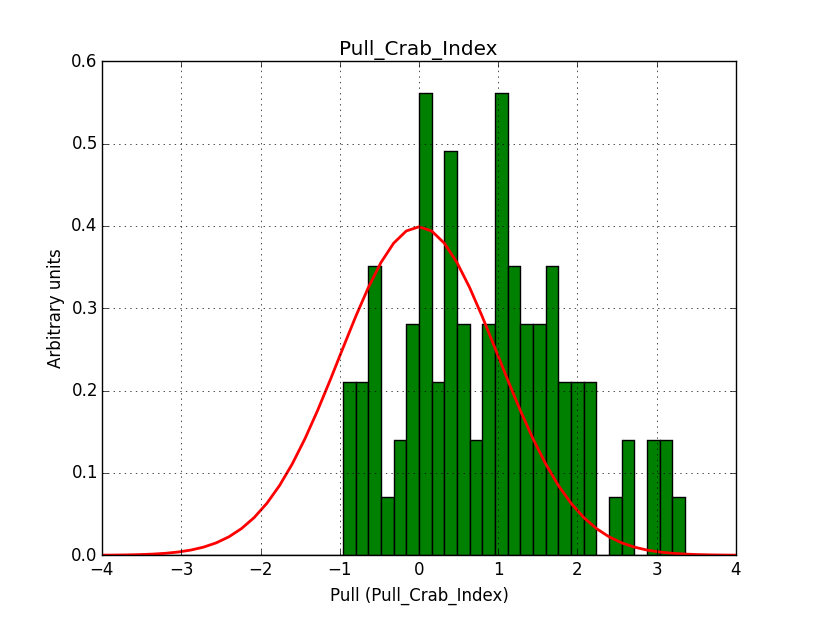
A bias occurs if the binning used is not sufficiently fine grained. In the actual simulations, a binning of 0.05 deg / pixel was used. This is slightly worse than the best angular resolution and introduces a biases, in particular in the fitted spectral index.
Here for reference the results for a 0.05 deg / pixel spatial binning: 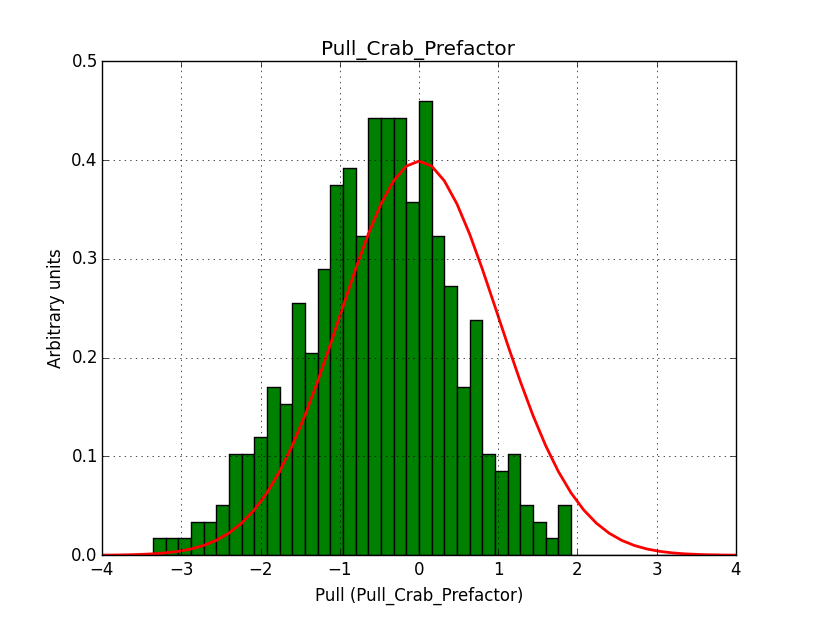 |
 |
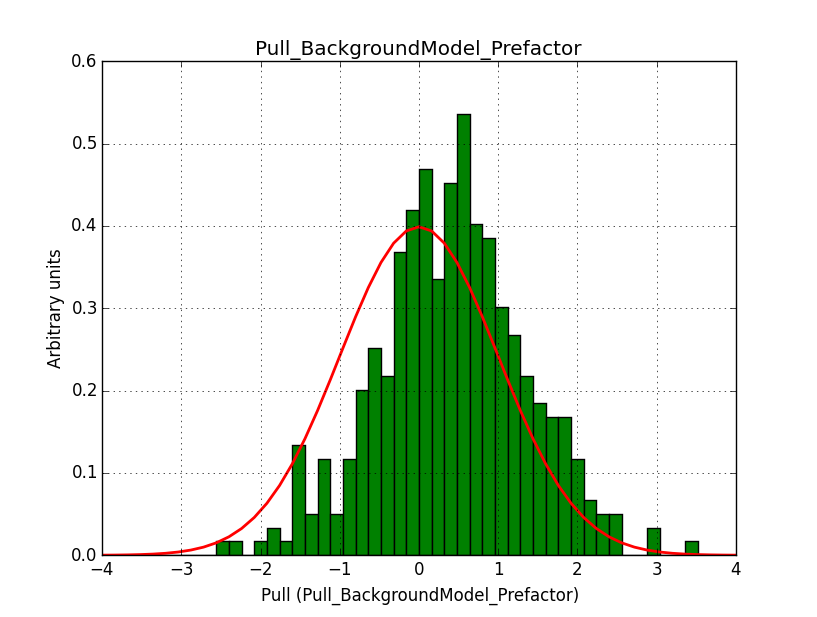 |
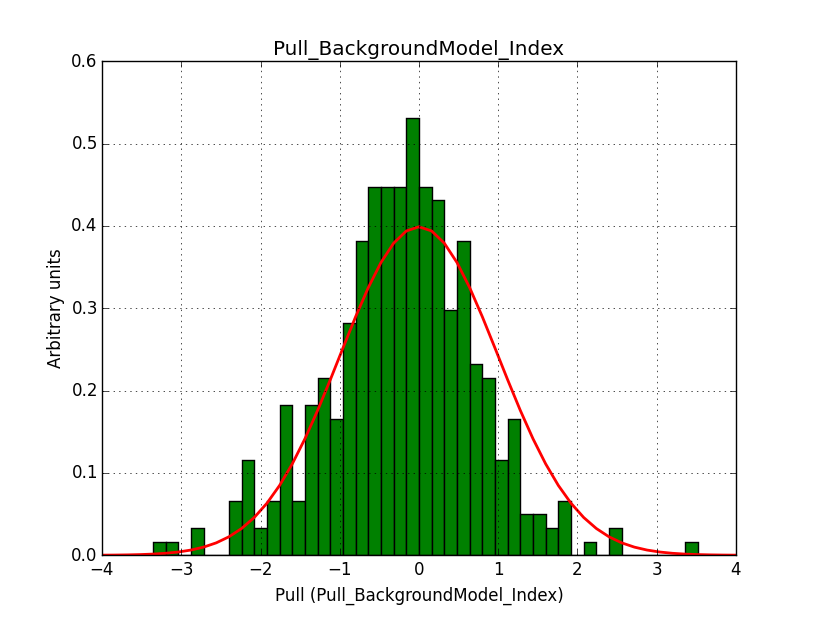 |
 |
 |
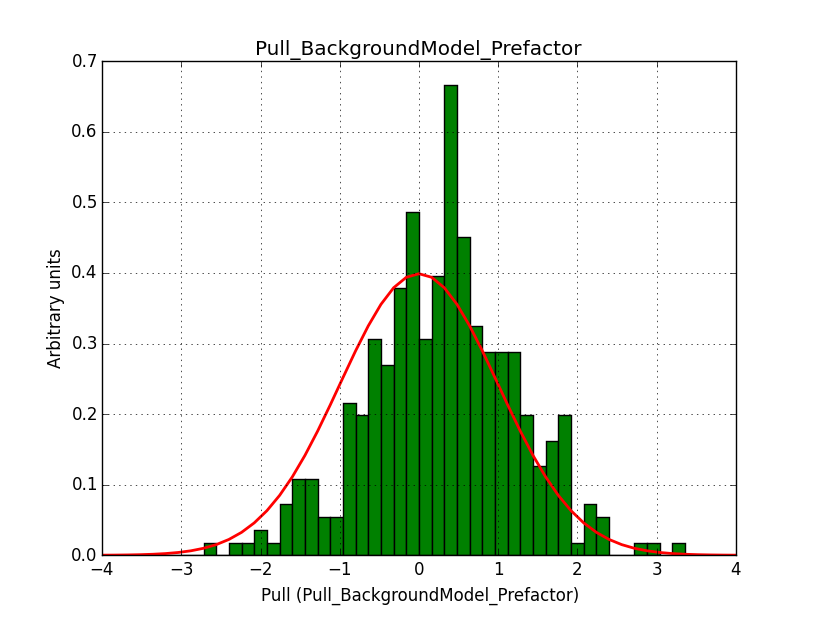 |
 |
#14
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- File new_binsz005_b20_bkg_index.png added
- File new_binsz005_b20_bkg_prefactor.png added
- File new_binsz005_b20_index.png added
- File new_binsz005_b20_prefactor.png added
#15
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- File new_binsz002_b20_bkg_index.png added
- File new_binsz002_b20_bkg_prefactor.png added
- File new_binsz002_b20_index.png added
- File new_binsz002_b20_prefactor.png added
#16
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- Status changed from In Progress to Feedback
- % Done changed from 90 to 100
I put this issue now on feedback.
We still need to do more pull distributions. I plan to do one for all spectral laws so that we also have a reference for the stacked analysis.
#17
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- File plaw_binsz002_b10_index.png added
- File plaw_binsz002_b10_prefactor.png added
- File plaw_binsz002_b15_index.png added
- File plaw_binsz002_b15_prefactor.png added
- File plaw_binsz002_b20_index.png added
- File plaw_binsz002_b20_prefactor.png added
- File plaw_binsz002_b30_index.png added
- File plaw_binsz002_b30_prefactor.png added
- File plaw_binsz002_b40_index.png added
- File plaw_binsz002_b40_prefactor.png added
Here some more results for 0.02° spatial binning an 10, 15, 20, 30 and 40 energy bins between 0.1 and 100 TeV. At least 30 energy bins are needed to reduce the bias to an acceptable level.
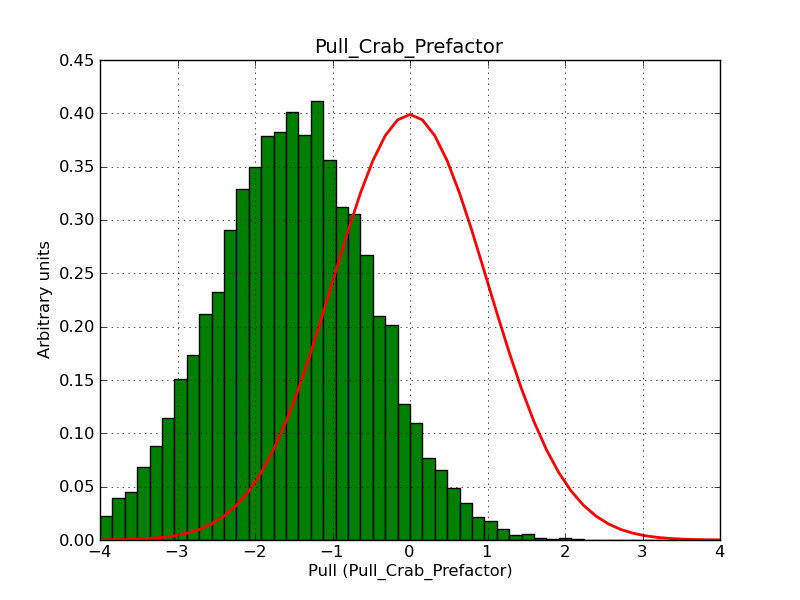 |
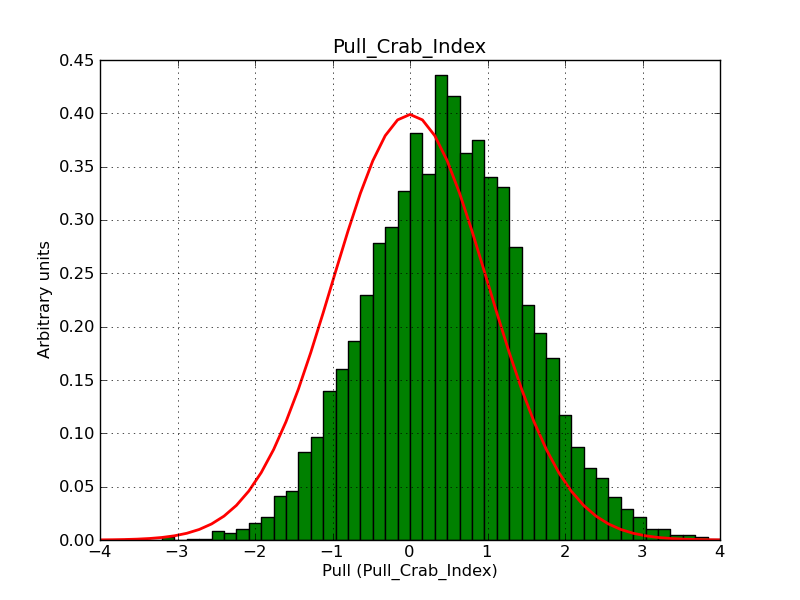 |
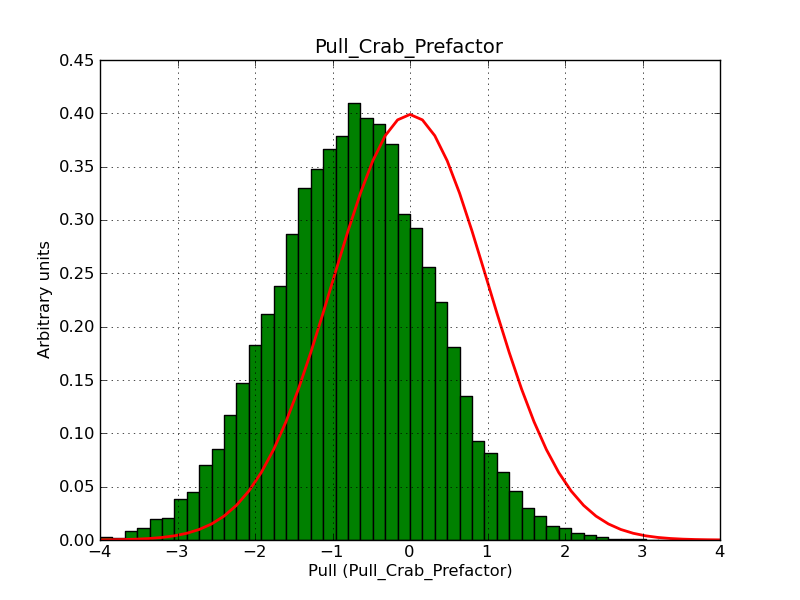 |
 |
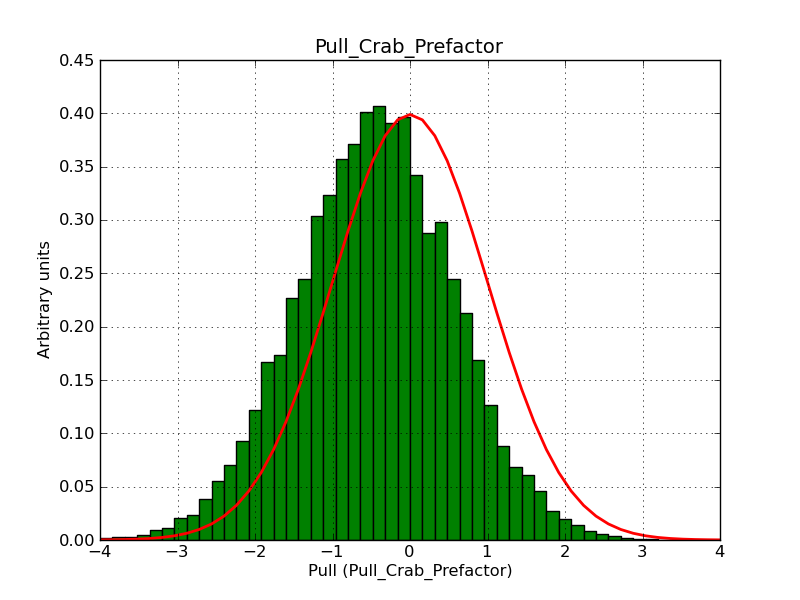 |
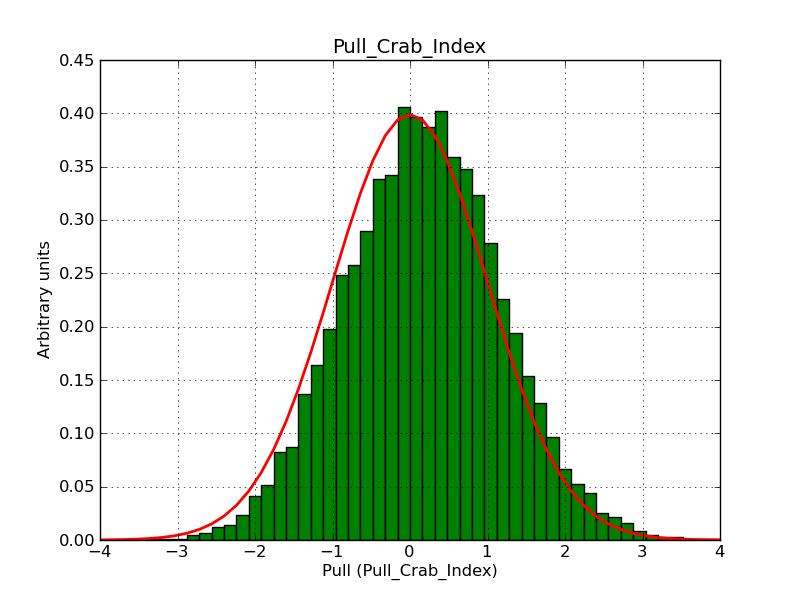 |
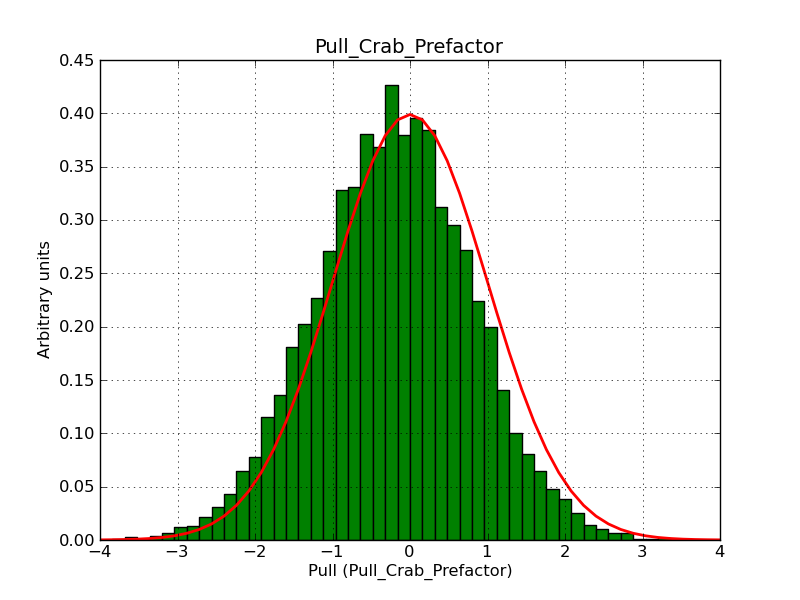 |
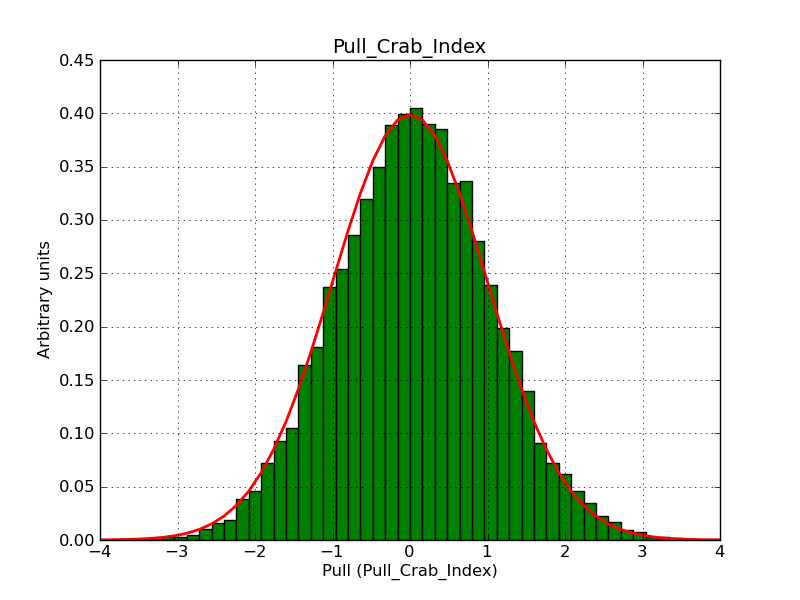 |
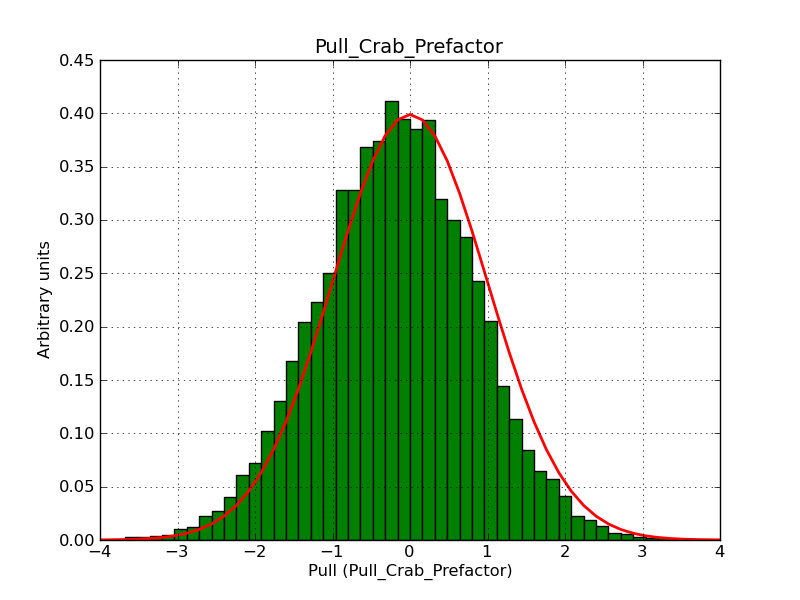 |
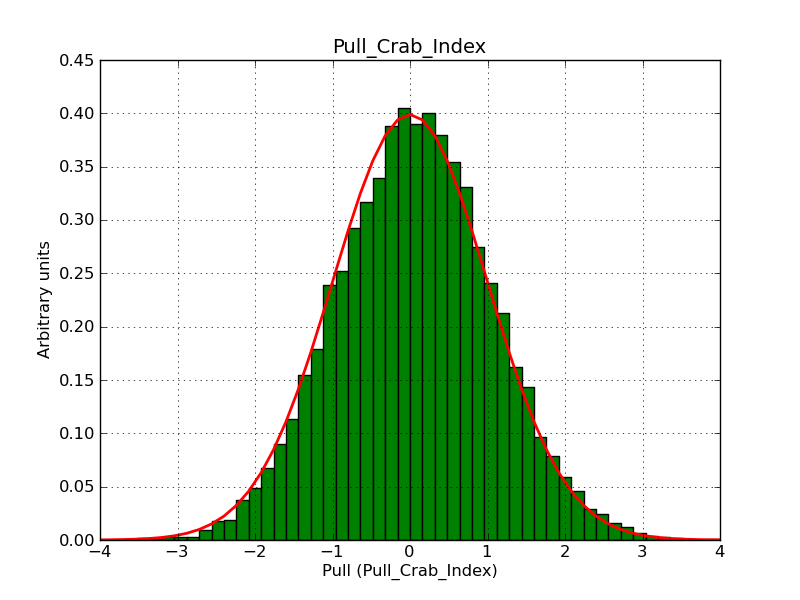 |
#18
 Updated by Knödlseder Jürgen over 9 years ago
Updated by Knödlseder Jürgen over 9 years ago
- Status changed from Feedback to Closed
Close this now as the issue was related to a limited number of bins in the initial runs that suggested a problem.